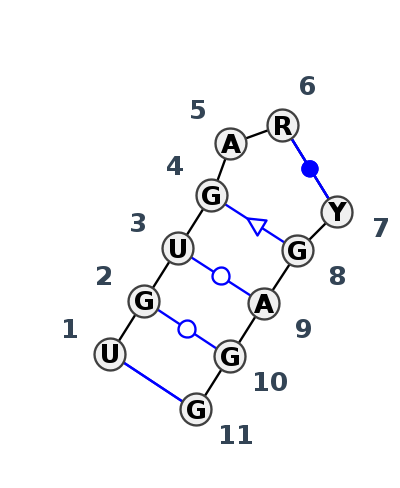
Motif IL_14684.1 Version IL_14684.1 of this group appears in releases 4.0 to 4.0
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | break | 7 | 8 | 9 | 10 | 11 | 1-11 | 2-10 | 3-9 | 4-8 | 6-7 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_8OI5_085 | 8OI5 | 0.0627 | 3 | 1 | LSU rRNA | U | 2151 | G | 2152 | U | 2154 | G | 2155 | A | 2156 | A | 2158 | * | U | 2134 | G | 2135 | A | 2136 | G | 2138 | G | 2139 | cWW | tWW | tWW | tSS | cWW | |
| 2 | IL_8GLP_114 | 8GLP | 0.0000 | 3 | Kink-turn related (H) | L5 | LSU rRNA | U | 3677 | G | 3678 | U | 3680 | G | 3681 | A | 3682 | G | 3684 | * | C | 3660 | G | 3661 | A | 3662 | G | 3664 | G | 3665 | cWW | tWW | tWW | tSS | cWW |
| 3 | IL_9AXU_079 | 9AXU | 0.0435 | 3 | 2 | LSU rRNA | U | 2261 | G | 2262 | U | 2264 | G | 2265 | A | 2266 | G | 2268 | * | C | 2244 | G | 2245 | A | 2246 | G | 2248 | G | 2249 | cWW | tWW | tWW | tSS | cWW | |
| 4 | IL_8P9A_311 | 8P9A | 0.0515 | 3 | Kink-turn related (H) | AR | LSU rRNA | U | 2173 | G | 2174 | U | 2176 | G | 2177 | A | 2178 | G | 2180 | * | C | 2156 | G | 2157 | A | 2158 | G | 2160 | G | 2161 | cWW | tWW | tWW | tSS | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| UGUUGACG*CGAUGG | 2 |
| UGUUGACA*UGAUGG | 1 |
| UGUUGACG*CGAAGG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GUUGAC*GAUG | 3 |
| GUUGAC*GAAG | 1 |
Release history
| Release | 4.0 |
|---|---|
| Date | 2025-08-13 |
| Status | New id, 1 parent |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- Kink-turn related (H) (2)
- (2)
- Basepair signature
- cWW-tWW-tWW-tSS-L-cWW
- Heat map statistics
- Min 0.04 | Avg 0.05 | Max 0.09
Coloring options: