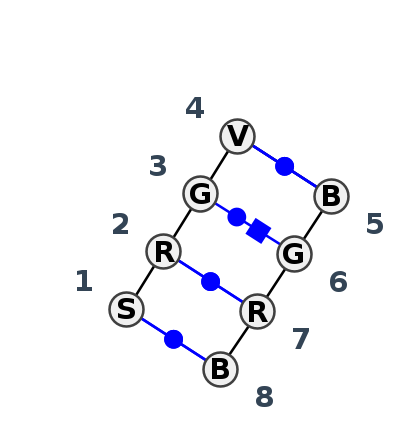
Motif IL_25872.3 Version IL_25872.3 of this group appears in releases 3.91 to 3.94
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | break | 5 | 6 | 7 | 8 | 1-8 | 2-7 | 3-6 | 4-5 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_7WIE_001 | 7WIE | 0.2270 | 0 | V | THF-II riboswitch (folE RNA) | G | 49 | A | 50 | G | 51 | A | 52 | * | U | 10 | G | 11 | G | 12 | U | 13 | cWW | cWW | cWH | cWW |
| 2 | IL_1DUQ_001 | 1DUQ | 0.0000 | 1 | A+B | THE REV BINDING ELEMENT | G | 104 | G | 105 | G | 106 | C | 107 | * | G | 123 | G | 124 | A | 126 | C | 127 | cWW | cWW | cWH | cWW |
| 3 | IL_1CSL_001 | 1CSL | 0.0578 | 1 | A+B | RNA (13-mer) + RNA (15-mer) | G | 46 | G | 47 | G | 48 | C | 49 | * | G | 70 | G | 71 | A | 73 | C | 74 | cWW | cWW | cWH | cWW |
| 4 | IL_7R9F_001 | 7R9F | 0.2411 | 0 | B | RNA (18-mer) | C | 5 | G | 6 | G | 7 | G | 8 | * | C | 5||||8_555 | G | 6||||8_555 | G | 7||||8_555 | G | 8||||8_555 | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GGGC*GGUAC | 2 |
| GAGA*UGGU | 1 |
| CGGG*CGGG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GG*GUA | 2 |
| AG*GG | 1 |
| GG*GG | 1 |
- Annotations
-
- (4)
- Basepair signature
- cWW-cWW-cWH-cWW
- Heat map statistics
- Min 0.06 | Avg 0.16 | Max 0.28
Coloring options: