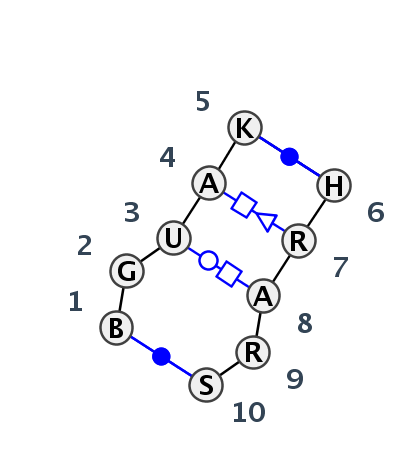
Motif IL_38862.1 Version IL_38862.1 of this group appears in releases 3.77 to 3.86
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | break | 6 | 7 | 8 | 9 | 10 | 1-10 | 2-3 | 2-9 | 3-8 | 4-7 | 5-6 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_2QWY_007 | 2QWY | 0.0000 | 0 | tSH-tWH-tHS | C | SAM-II riboswitch | C | 16 | G | 17 | U | 18 | A | 19 | U | 20 | * | A | 46 | A | 47 | A | 48 | A | 49 | G | 50 | cWW | tSH | tWH | tHS | cWW | |
| 2 | IL_5J7L_300 | 5J7L | 0.2806 | 0 | tSH-tWH-tHS | DA | LSU rRNA | U | 1474 | G | 1475 | U | 1476 | A | 1477 | G | 1478 | * | U | 1513 | G | 1514 | A | 1515 | G | 1516 | G | 1517 | cWW | tSH | tWH | tHS | cWW | |
| 3 | IL_7RQB_027 | 7RQB | 0.5110 | 0 | 5x5 Sarcin-Ricin with intercalated A; G-bulge | 1A | LSU rRNA | U | 858 | G | 859 | U | 860 | A | 861 | G | 862 | * | C | 915 | G | 916 | A | 917 | A | 918 | G | 919 | cWW | cSH | tWH | tHS | cWW | |
| 4 | IL_4V9F_033 | 4V9F | 0.5078 | 0 | 5x5 Sarcin-Ricin with intercalated A; G-bulge | 0 | LSU rRNA | G | 952 | G | 953 | U | 954 | A | 955 | G | 956 | * | C | 1011 | A | 1012 | A | 1013 | A | 1014 | C | 1015 | cWW | cSH | tWH | tHS | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CGUAU*AAAAG | 1 |
| UGUAG*UGAGG | 1 |
| UGUAG*CGAAG | 1 |
| GGUAG*CAAAC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GUA*AAA | 2 |
| GUA*GAG | 1 |
| GUA*GAA | 1 |
Release history
| Release | 3.77 | 3.78 | 3.79 | 3.80 | 3.81 | 3.82 | 3.83 | 3.84 | 3.85 | 3.86 |
|---|---|---|---|---|---|---|---|---|---|---|
| Date | 2023-12-06 | 2024-01-03 | 2024-01-03 | 2024-01-31 | 2024-02-28 | 2024-03-27 | 2024-04-24 | 2024-05-22 | 2024-06-19 | 2024-07-17 |
| Status | New id, 2 parents | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- tSH-tWH-tHS (2)
- 5x5 Sarcin-Ricin with intercalated A; G-bulge (2)
- Basepair signature
- cWW-L-R-tWH-tHS-cWW
- Heat map statistics
- Min 0.11 | Avg 0.29 | Max 0.51
Coloring options: