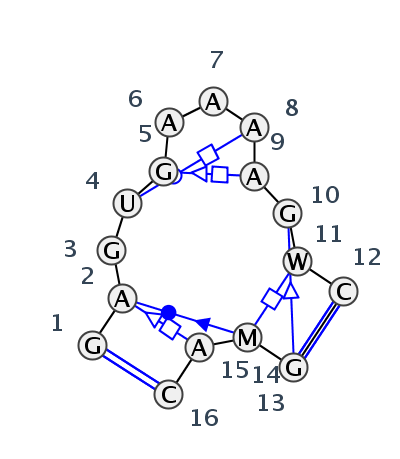
Motif IL_60992.5 Version IL_60992.5 of this group appears in releases 3.54 to 3.79
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | break | 13 | 14 | 15 | 16 | 1-16 | 2-14 | 2-15 | 3-10 | 4-8 | 5-9 | 10-13 | 11-14 | 12-13 | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_7A0S_009 | 7A0S | 0.1057 | 2 | X | LSU rRNA | G | 506 | A | 507 | G | 508 | U | 509 | G | 510 | A | 511 | A | 512 | A | 513 | A | 515 | G | 516 | A | 517 | C | 519 | * | G | 492 | A | 493 | A | 494 | C | 495 | cWW | cWS | tSH | cWB | tWH | ntSH | tSS | ntHH | cWW |
| 2 | IL_5J7L_255 | 5J7L | 0.0000 | 2 | DA | LSU rRNA | G | 496 | A | 497 | G | 498 | U | 499 | G | 500 | A | 501 | A | 502 | A | 503 | A | 505 | G | 506 | A | 507 | C | 509 | * | G | 481 | A | 482 | A | 483 | C | 484 | cWW | cWS | tSH | cWB | tWH | tSH | tSS | ntHH | cWW |
| 3 | IL_7RQB_014 | 7RQB | 0.1249 | 2 | 1A | LSU rRNA | G | 496 | A | 497 | G | 498 | U | 499 | G | 500 | A | 501 | A | 502 | A | 503 | A | 505 | G | 506 | A | 507 | C | 509 | * | G | 481 | A | 482 | A | 483 | C | 484 | cWW | cWS | tSH | cWB | tWH | tSH | tSS | ntHH | cWW |
| 4 | IL_4WF9_015 | 4WF9 | 0.1298 | 2 | X | LSU rRNA | G | 541 | A | 542 | G | 543 | U | 544 | G | 545 | A | 546 | A | 547 | A | 548 | A | 550 | G | 551 | A | 552 | C | 554 | * | G | 527 | C | 528 | A | 529 | C | 530 | cWW | cWS | tSH | cWB | tWH | ntSH | tSS | cWW | |
| 5 | IL_5TBW_014 | 5TBW | 0.2494 | 2 | 1 | LSU rRNA | G | 390 | A | 391 | G | 392 | U | 393 | G | 394 | A | 395 | A | 396 | A | 397 | A | 399 | G | 400 | U | 401 | C | 403 | * | G | 376 | A | 377 | A | 378 | C | 379 | cWW | ncWS | tSH | cWB | tWH | tSH | tSS | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GAGUGAAAGAGAAC*GAAC | 1 |
| GAGUGAAAAAGAAC*GAAC | 1 |
| GAGUGAAAUAGAGC*GAAC | 1 |
| GAGUGAAAUAGAAC*GCAC | 1 |
| GAGUGAAAAAGUAC*GAAC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| AGUGAAAGAGAA*AA | 1 |
| AGUGAAAAAGAA*AA | 1 |
| AGUGAAAUAGAG*AA | 1 |
| AGUGAAAUAGAA*CA | 1 |
| AGUGAAAAAGUA*AA | 1 |
Release history
| Release | 3.54 | 3.55 | 3.56 | 3.57 | 3.58 | 3.59 | 3.60 | 3.61 | 3.62 | 3.63 | 3.64 | 3.65 | 3.66 | 3.67 | 3.68 | 3.69 | 3.70 | 3.71 | 3.72 | 3.73 | 3.74 | 3.75 | 3.76 | 3.77 | 3.78 | 3.79 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Date | 2022-02-02 | 2022-03-02 | 2022-03-30 | 2022-04-27 | 2022-05-25 | 2022-06-22 | 2022-07-20 | 2022-08-17 | 2022-10-12 | 2022-11-09 | 2022-12-07 | 2023-01-04 | 2023-02-01 | 2023-03-01 | 2023-03-29 | 2023-04-26 | 2023-05-24 | 2023-06-21 | 2023-07-19 | 2023-08-16 | 2023-09-13 | 2023-10-11 | 2023-11-08 | 2023-12-06 | 2024-01-03 | 2024-01-03 |
| Status | Updated, 1 parent | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (5)
- Basepair signature
- cWW-tSS-tHH-cSW-bif-tSH-tWH-R-R-tHS-cWW
- Heat map statistics
- Min 0.11 | Avg 0.15 | Max 0.30
Coloring options: