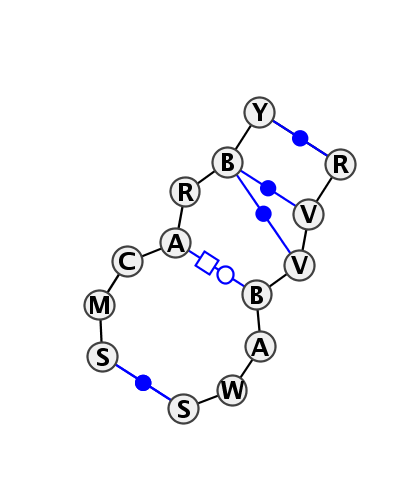
Motif IL_65851.1 Version IL_65851.1 of this group appears in releases 3.62 to 3.76
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | break | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 1-14 | 2-13 | 3-11 | 3-12 | 3-13 | 4-11 | 5-10 | 6-9 | 6-10 | 7-8 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_6VMY_004 | 6VMY | 0.7097 | 1 | A | Cobalamin riboswitch | C | 326 | A | 327 | C | 328 | A | 330 | A | 331 | U | 332 | U | 333 | * | A | 342 | C | 343 | A | 344 | G | 345 | A | 346 | U | 347 | G | 348 | cWW | ntHH | ncWW | ncWW | cWW | ||||||
| 2 | IL_6CZR_121 | 6CZR | 0.0000 | 1 | tWH-tWH-tHW-tHW | 1a | SSU rRNA | G | 143 | A | 144 | C | 145 | A | 146 | A | 147 | C | 148 | C | 149 | * | G | 162 | G | 163 | C | 164 | U | 165 | A | 166 | A | 167 | C | 169 | cWW | ncWB | ntWH | ntHW | ncWB | ncWW | ||||
| 3 | IL_3NDB_008 | 3NDB | 0.5975 | 0 | M | SRP RNA | G | 200 | C | 201 | C | 202 | A | 203 | G | 204 | G | 205 | C | 206 | * | G | 215 | A | 216 | G | 217 | C | 218 | A | 219 | A | 220 | C | 221 | cWW | cWW | ntWH | ntSH | tHW | tHS | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CACGAAUU*ACAGAUG | 1 |
| GACAACC*GGCUAAUC | 1 |
| GCCAGGC*GAGCAAC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| ACGAAU*CAGAU | 1 |
| ACAAC*GCUAAU | 1 |
| CCAGG*AGCAA | 1 |
Release history
| Release | 3.62 | 3.63 | 3.64 | 3.65 | 3.66 | 3.67 | 3.68 | 3.69 | 3.70 | 3.71 | 3.72 | 3.73 | 3.74 | 3.75 | 3.76 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Date | 2022-10-12 | 2022-11-09 | 2022-12-07 | 2023-01-04 | 2023-02-01 | 2023-03-01 | 2023-03-29 | 2023-04-26 | 2023-05-24 | 2023-06-21 | 2023-07-19 | 2023-08-16 | 2023-09-13 | 2023-10-11 | 2023-11-08 |
| Status | New id, > 2 parents | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (2)
- tWH-tWH-tHW-tHW (1)
- Basepair signature
- cWW-L-R-R-L-tHW-L-cWW-cWW-cWW
- Heat map statistics
- Min 0.60 | Avg 0.47 | Max 0.82
Coloring options: