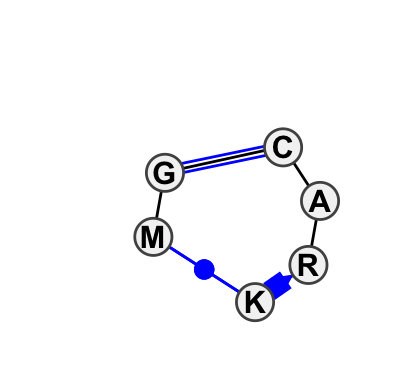
Motif IL_66718.1 Version IL_66718.1 of this group appears in releases 0.5 to 0.5
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | break | 3 | 4 | 5 | 6 | 1-6 | 2-3 | 4-6 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_3NPB_005 | 3NPB | 0.0000 | 0 | A | SAM-I riboswitch | C | 49 | G | 50 | * | C | 73 | A | 74 | A | 75 | G | 76 | cWW | cWW | ncSH |
| 2 | IL_1S72_009 | 1S72 | 0.8369 | 2 | 0 | LSU rRNA | A | 248 | G | 249 | * | C | 260 | A | 261 | G | 264 | U | 265 | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CG*CAAG | 1 |
| AG*CAAUGU | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| *AA | 1 |
| *AAUG | 1 |
Release history
| Release | 0.5 |
|---|---|
| Date | 2011-12-08 |
| Status | New id, 1 parent |
Parent motifs
| Parent motif | Common motif instances | Only in IL_66718.1 | Only in the parent motif |
|---|---|---|---|
| IL_50503.1 Compare | IL_1S72_009 | IL_3NPB_005 |
Children motifs
| Child motif | Common motif instances | Only in IL_66718.1 | Only in the child motif |
|---|---|---|---|
| IL_92484.1 Compare | IL_1S72_009 | IL_3NPB_005 | IL_4A1B_004, IL_3U5H_007, IL_2XZM_018, IL_2XZM_062 |
| IL_78809.1 Compare | IL_3NPB_005 | IL_1S72_009 |
Coloring options: