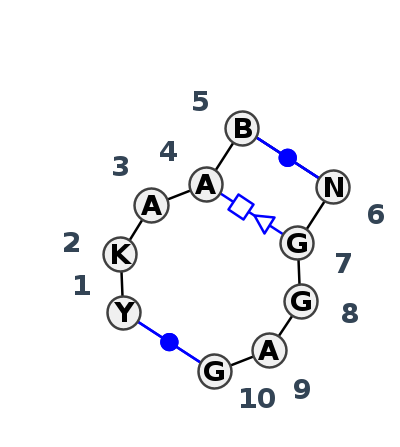
Motif IL_66798.1 Version IL_66798.1 of this group appears in releases 3.89 to 3.90
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | break | 6 | 7 | 8 | 9 | 10 | 1-10 | 2-9 | 2-10 | 3-8 | 4-7 | 5-6 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_4WF9_019 | 4WF9 | 0.1579 | 0 | AAA cross-strand stack | X | LSU rRNA | U | 649 | U | 650 | A | 651 | A | 652 | G | 653 | * | U | 663 | G | 664 | G | 665 | A | 666 | G | 667 | cWW | ntWW | tHS | cWW | ||
| 2 | IL_7A0S_013 | 7A0S | 0.1160 | 0 | AAA cross-strand stack | X | LSU rRNA | U | 616 | U | 617 | A | 618 | A | 619 | G | 620 | * | C | 629 | G | 630 | G | 631 | A | 632 | G | 633 | cWW | tHS | cWW | |||
| 3 | IL_8VTW_015 | 8VTW | 0.0000 | 0 | AAA cross-strand stack (H) | 1A | LSU rRNA | U | 606 | U | 607 | A | 608 | A | 609 | G | 610 | * | C | 618 | G | 619 | G | 620 | A | 621 | G | 622 | cWW | ntWH | tHS | cWW | ||
| 4 | IL_5J7L_258 | 5J7L | 0.1089 | 0 | AAA cross-strand stack | DA | LSU rRNA | U | 606 | U | 607 | A | 608 | A | 609 | C | 610 | * | G | 618 | G | 619 | G | 620 | A | 621 | G | 622 | cWW | ntWH | tHS | cWW | ||
| 5 | IL_4V9F_020 | 4V9F | 0.1629 | 0 | AAA cross-strand stack | 0 | LSU rRNA | C | 663 | U | 664 | A | 665 | A | 666 | C | 667 | * | G | 679 | G | 680 | G | 681 | A | 682 | G | 683 | cWW | ntWH | tHS | cWW | ||
| 6 | IL_4WF9_074 | 4WF9 | 0.5254 | 0 | Triple sheared | X | LSU rRNA | C | 1909 | G | 1910 | A | 1911 | A | 1912 | U | 1913 | * | A | 1883 | G | 1884 | G | 1885 | A | 1886 | G | 1887 | ncWW | cSW | tHS | ntHS | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| UUAAG*CGGAG | 2 |
| UUAAG*UGGAG | 1 |
| UUAAC*GGGAG | 1 |
| CUAAC*GGGAG | 1 |
| CGAAU*AGGAG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| UAA*GGA | 5 |
| GAA*GGA | 1 |
- Annotations
-
- AAA cross-strand stack (4)
- Triple sheared (1)
- AAA cross-strand stack (H) (1)
- Basepair signature
- cWW-L-R-L-R-tHS-cWW
- Heat map statistics
- Min 0.11 | Avg 0.25 | Max 0.59
Coloring options: