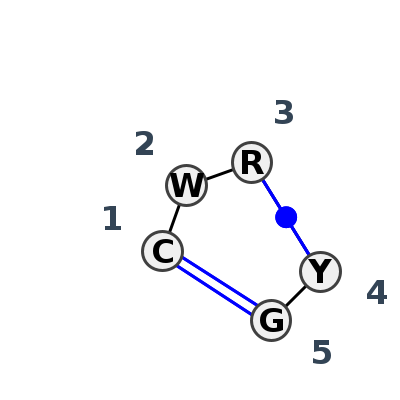
Motif IL_75501.1 Version IL_75501.1 of this group appears in releases 4.1 to 4.4
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | break | 4 | 5 | 1-5 | 3-4 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_8GLP_036 | 8GLP | 0.4198 | 2 | L5 | LSU rRNA | C | 727 | U | 728 | G | 731 | * | C | 938 | G | 939 | cWW | cWW | |
| 2 | IL_2QWY_008 | 2QWY | 0.0000 | 0 | Single stack bend with SAM intercalation | C | SAM-II riboswitch | C | 43 | U | 44 | A | 45 | * | U | 21 | G | 22 | cWW | cWW |
| 3 | IL_6FZ0_001 | 6FZ0 | 0.1076 | 0 | Single stack bend with SAM intercalation | A | SAM-V riboswitch | C | 46 | U | 47 | A | 48 | * | U | 20 | G | 21 | cWW | cWW |
| 4 | IL_3MOJ_004 | 3MOJ | 0.4497 | 0 | Single stack bend | A | LSU rRNA | C | 2540 | A | 2541 | A | 2542 | * | U | 2522 | G | 2523 | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CUA*UG | 2 |
| CUGGG*CG | 1 |
| CAA*UG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| U* | 2 |
| UGG* | 1 |
| A* | 1 |
- Annotations
-
- Single stack bend with SAM intercalation (2)
- Single stack bend (1)
- (1)
- Basepair signature
- cWW-L-cWW
- Heat map statistics
- Min 0.11 | Avg 0.32 | Max 0.69
Coloring options: