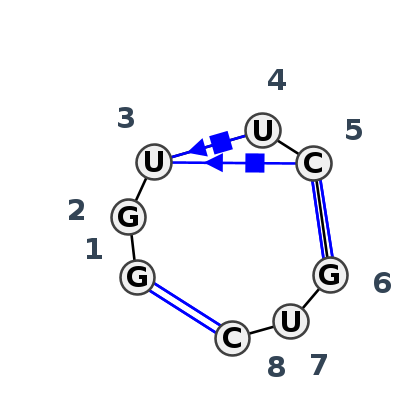
Motif IL_83497.1 Version IL_83497.1 of this group appears in releases 4.1 to 4.6
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | break | 6 | 7 | 8 | 1-8 | 2-7 | 3-4 | 3-5 | 4-7 | 5-6 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_6DLR_002 | 6DLR | 0.1234 | 0 | 8-nt loop receptor | A | Guanidine-I riboswitch | G | 16 | G | 17 | U | 18 | U | 19 | C | 20 | * | G | 42 | U | 43 | C | 44 | cWW | cWB | ncSH | cSH | ncWw | cWW |
| 2 | IL_7MLW_003 | 7MLW | 0.1213 | 0 | F | Guanidine-I riboswitch | G | 11 | G | 12 | U | 13 | U | 14 | C | 15 | * | G | 48 | U | 49 | C | 50 | cWW | cWB | cSH | cSH | cWW | ||
| 3 | IL_5U3G_002 | 5U3G | 0.0000 | 0 | 8-nt loop receptor | B | Guanidine-I riboswitch | G | 10 | G | 11 | U | 12 | U | 13 | C | 14 | * | G | 27 | U | 28 | C | 29 | cWW | cWB | cSH | ncWw | cWW | |
| 4 | IL_5T83_002 | 5T83 | 0.1022 | 0 | 8-nt loop receptor | A | Guanidine-I riboswitch | G | 16 | G | 17 | U | 18 | U | 19 | C | 20 | * | G | 39 | U | 40 | C | 41 | cWW | ncWB | cSH | ncSH | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GGUUC*GUC | 4 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GUU*U | 4 |
- Annotations
-
- 8-nt loop receptor (3)
- (1)
- Basepair signature
- cWW-L-R-cSH-cSH-cWW
- Heat map statistics
- Min 0.10 | Avg 0.09 | Max 0.13
Coloring options: