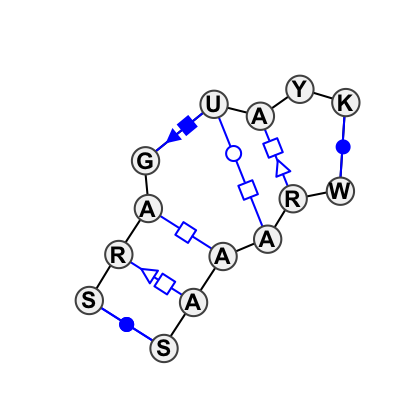
Motif IL_95652.5 Version IL_95652.5 of this group appears in releases 1.17 to 1.17
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | break | 9 | 10 | 11 | 12 | 13 | 14 | 1-14 | 2-13 | 3-12 | 4-5 | 5-11 | 6-10 | 8-9 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_2AW7_041 | 2AW7 | 0.1378 | 0 | A | SSU rRNA | G | 887 | G | 888 | A | 889 | G | 890 | U | 891 | A | 892 | C | 893 | G | 894 | * | U | 905 | A | 906 | A | 907 | A | 908 | A | 909 | C | 910 | cWW | tSH | tHH | cSH | tWH | tHS | cWW | |
| 2 | IL_1FJG_034 | 1FJG | 0.1180 | 0 | 8x6 Sarcin-Ricin with inserted Y; G-bulge (H) | A | SSU rRNA | G | 887 | G | 888 | A | 889 | G | 890 | U | 891 | A | 892 | C | 893 | G | 894 | * | U | 905 | G | 906 | A | 907 | A | 908 | A | 909 | C | 910 | cWW | tSH | tHH | cSH | tWH | tHS | cWW |
| 3 | IL_3U5F_057 | 3U5F | 0.0976 | 0 | 6 | SSU rRNA | G | 1111 | G | 1112 | A | 1113 | G | 1114 | U | 1115 | A | 1116 | U | 1117 | G | 1118 | * | U | 1129 | G | 1130 | A | 1131 | A | 1132 | A | 1133 | C | 1134 | cWW | tSH | tHH | cSH | tWH | tHS | cWW | |
| 4 | IL_4BPP_052 | 4BPP | 0.0000 | 0 | A | SSU rRNA | G | 1083 | A | 1084 | A | 1085 | G | 1086 | U | 1087 | A | 1088 | U | 1089 | G | 1090 | * | U | 1102 | G | 1103 | A | 1104 | A | 1105 | A | 1106 | C | 1107 | cWW | tSH | tHH | cSH | tWH | tHS | cWW | |
| 5 | IL_4CUX_055 | 4CUX | 0.1071 | 0 | 2 | SSU rRNA | G | 1111 | G | 1112 | A | 1113 | G | 1114 | U | 1115 | A | 1116 | U | 1117 | G | 1118 | * | U | 1129 | G | 1130 | A | 1131 | A | 1132 | A | 1133 | C | 1134 | cWW | tSH | ntHH | ntWH | tHS | cWW | ||
| 6 | IL_3J7A_050 | 3J7A | 0.1566 | 0 | 8x6 Sarcin-Ricin with inserted Y; G-bulge (H) | A | SSU rRNA | C | 1212 | G | 1213 | A | 1214 | G | 1215 | U | 1216 | A | 1217 | U | 1218 | U | 1219 | * | A | 1230 | G | 1231 | A | 1232 | A | 1233 | A | 1234 | G | 1235 | cWW | tSH | tHH | cSH | tWH | tHS | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GGAGUAUG*UGAAAC | 2 |
| GGAGUACG*UAAAAC | 1 |
| GGAGUACG*UGAAAC | 1 |
| GAAGUAUG*UGAAAC | 1 |
| CGAGUAUU*AGAAAG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GAGUAU*GAAA | 3 |
| GAGUAC*AAAA | 1 |
| GAGUAC*GAAA | 1 |
| AAGUAU*GAAA | 1 |
Release history
| Release | 1.17 |
|---|---|
| Date | 2014-09-04 |
| Status | Updated, 1 parent |
Parent motifs
| Parent motif | Common motif instances | Only in IL_95652.5 | Only in the parent motif |
|---|---|---|---|
| IL_95652.4 Compare | IL_2AW7_041, IL_3U5F_057, IL_4BPP_052, IL_1FJG_034, IL_4CUX_055 | IL_3J7A_050 |
Children motifs
| Child motif | Common motif instances | Only in IL_95652.5 | Only in the child motif |
|---|---|---|---|
| IL_95652.6 Compare | IL_2AW7_041, IL_1FJG_034, IL_3J7A_050, IL_4CUX_055, IL_3U5F_057, IL_4BPP_052 | IL_4W23_083 |
- Annotations
-
- (4)
- 8x6 Sarcin-Ricin with inserted Y; G-bulge (H) (2)
- Basepair signature
- cWW-tSH-tHH-cSH-tWH-tHS-L-cWW
- Heat map statistics
- Min 0.10 | Avg 0.12 | Max 0.20
Coloring options: