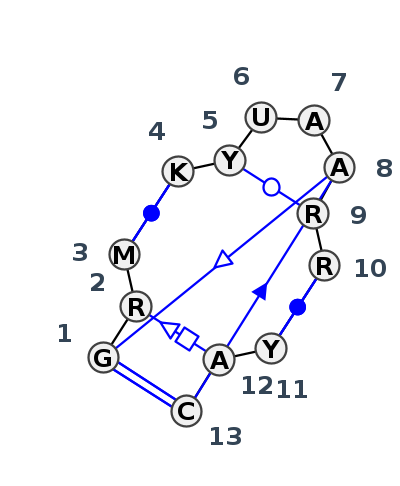
Motif J3_28621.5 Version J3_28621.5 of this group appears in releases 3.95 to 3.96
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name | 1 | 2 | 3 | break | 4 | 5 | 6 | 7 | 8 | 9 | 10 | break | 11 | 12 | 13 | 1-8 | 1-13 | 2-12 | 3-4 | 5-9 | 8-13 | 10-11 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | J3_5J7L_040 | 5J7L | 0.1257 | 0 | DA | LSU rRNA | G | 1055 | G | 1056 | A | 1057 | * | U | 1081 | U | 1082 | U | 1083 | A | 1084 | A | 1085 | A | 1086 | G | 1087 | * | C | 1102 | A | 1103 | C | 1104 | tsS | cWW | tSH | cWW | tWW | cSs | cWW |
| 2 | J3_5D8H_001 | 5D8H | 0.0000 | 0 | A | LSU rRNA | G | 1165 | G | 1166 | A | 1167 | * | U | 1191 | U | 1192 | U | 1193 | A | 1194 | A | 1195 | A | 1196 | G | 1197 | * | C | 1212 | A | 1213 | C | 1214 | tsS | cWW | tSH | cWW | tWW | ncSs | cWW |
| 3 | J3_1MMS_001 | 1MMS | 0.0898 | 0 | C | 23S RIBOSOMAL RNA | G | 1055 | G | 1056 | A | 1057 | * | U | 1081 | U | 1082 | U | 1083 | A | 1084 | A | 1085 | A | 1086 | G | 1087 | * | C | 1102 | A | 1103 | C | 1104 | tsS | cWW | tSH | cWW | tWW | ncSs | cWW |
| 4 | J3_6PRV_001 | 6PRV | 0.1098 | 0 | A | LSU rRNA | G | 1055 | G | 1056 | A | 1057 | * | U | 1081 | U | 1082 | U | 1083 | A | 1084 | A | 1085 | A | 1086 | G | 1087 | * | C | 1102 | A | 1103 | C | 1104 | tsS | cWW | tSH | cWW | tWW | cSs | cWW |
| 5 | J3_4V9F_004 | 4V9F | 0.1280 | 0 | 0 | LSU rRNA | G | 1159 | G | 1160 | A | 1161 | * | U | 1185 | C | 1186 | U | 1187 | A | 1188 | A | 1189 | G | 1190 | A | 1191 | * | U | 1206 | A | 1207 | C | 1208 | tsS | cWW | cWW | tWW | cWW | ||
| 6 | J3_8P9A_052 | 8P9A | 0.1543 | 0 | AR | LSU rRNA | G | 1230 | A | 1231 | C | 1232 | * | G | 1256 | C | 1257 | U | 1258 | A | 1259 | A | 1260 | G | 1261 | G | 1262 | * | C | 1277 | A | 1278 | C | 1279 | ntsS | cWW | tSH | cWW | ntWW | cSs | cWW |
| 7 | J3_8CRE_012 | 8CRE | 0.2139 | 0 | 1 | LSU rRNA | G | 1226 | A | 1227 | C | 1228 | * | G | 1252 | C | 1253 | U | 1254 | A | 1255 | A | 1256 | G | 1257 | G | 1258 | * | C | 1273 | A | 1274 | C | 1275 | ntSs | cWW | ntSH | cWW | tWW | ncSs | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GGA*UUUAAAG*CAC | 4 |
| GAC*GCUAAGG*CAC | 2 |
| GGA*UCUAAGA*UAC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| G*UUAAA*A | 4 |
| A*CUAAG*A | 2 |
| G*CUAAG*A | 1 |
- Annotations
-
- (7)
- Basepair signature
- cWW-tSS-cSS-tSH-cWW-cWW-tWW-F-F
- Heat map statistics
- Min 0.09 | Avg 0.14 | Max 0.23
Coloring options: