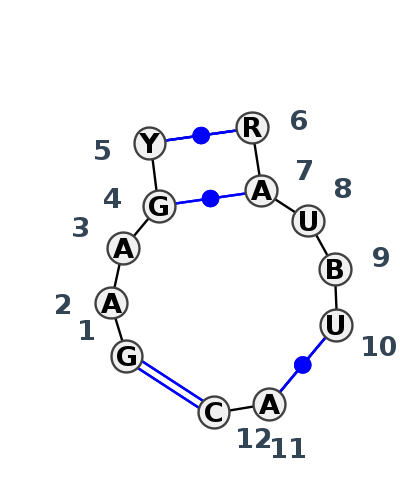
Motif J3_38265.2 Version J3_38265.2 of this group appears in releases 3.90 to 3.90
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | break | 6 | 7 | 8 | 9 | 10 | break | 11 | 12 | 1-2 | 1-12 | 4-7 | 5-6 | 10-11 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | J3_8C3A_060 | 8C3A | 0.1637 | 2 | 4 | 5.8S rRNA | G | 58 | A | 59 | A | 61 | G | 63 | U | 64 | * | A | 96 | A | 97 | U | 98 | C | 99 | U | 100 | * | A | 44 | C | 45 | ncSs | cWW | cWW | cWW | cWW |
| 2 | J3_8P4V_024 | 8P4V | 0.1346 | 2 | 4 | 5.8S rRNA | G | 58 | A | 59 | A | 61 | G | 63 | U | 64 | * | A | 96 | A | 97 | U | 98 | C | 99 | U | 100 | * | A | 44 | C | 45 | ncSs | cWW | cWW | cWW | cWW |
| 3 | J3_5TBW_071 | 5TBW | 0.1234 | 2 | 4 | 5.8S rRNA | G | 58 | A | 59 | A | 61 | G | 63 | U | 64 | * | A | 96 | A | 97 | U | 98 | C | 99 | U | 100 | * | A | 44 | C | 45 | ncSs | cWW | cWW | cWW | cWW |
| 4 | J3_5J7L_037 | 5J7L | 0.0571 | 2 | DA | LSU rRNA | G | 70 | A | 71 | A | 73 | G | 75 | C | 76 | * | G | 110 | A | 111 | U | 112 | U | 113 | U | 114 | * | A | 56 | C | 57 | ncSs | cWW | cWW | cWW | cWW |
| 5 | J3_4V9F_014 | 4V9F | 0.0000 | 2 | 0 | LSU rRNA | G | 66 | A | 67 | A | 69 | G | 71 | C | 72 | * | G | 105 | A | 106 | U | 107 | U | 108 | U | 109 | * | A | 52 | C | 53 | ncSs | cWW | cWW | cWW | cWW |
| 6 | J3_8VTW_039 | 8VTW | 0.0759 | 2 | 1A | LSU rRNA | G | 70 | A | 71 | A | 73 | G | 75 | C | 76 | * | G | 110 | A | 111 | U | 112 | G | 113 | U | 114 | * | A | 56 | C | 57 | ncSs | cWW | cWW | cWW | cWW |
| 7 | J3_7A0S_002 | 7A0S | 0.1461 | 2 | X | LSU rRNA | G | 69 | A | 70 | A | 72 | G | 74 | C | 75 | * | G | 108 | A | 109 | U | 110 | G | 111 | U | 112 | * | A | 55 | C | 56 | cWW | ncWW | cWW | cWW | |
| 8 | J3_4WF9_013 | 4WF9 | 0.1671 | 2 | X | LSU rRNA | G | 70 | A | 71 | A | 73 | G | 75 | C | 76 | * | G | 109 | A | 110 | U | 111 | U | 112 | U | 113 | * | A | 56 | C | 57 | cSH | cWW | cWW | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| AC*GAUACGU*AAUCU | 3 |
| AC*GAUAAGC*GAUUU | 2 |
| AC*GAUAAGC*GAUGU | 1 |
| AC*GAAAAGC*GAUGU | 1 |
| AC*GAUAUGC*GAUUU | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| *AUACG*AUC | 3 |
| *AUAAG*AUU | 2 |
| *AUAAG*AUG | 1 |
| *AAAAG*AUG | 1 |
| *AUAUG*AUU | 1 |
Release history
| Release | 3.90 |
|---|---|
| Date | 2024-11-06 |
| Status | Updated, 1 parent |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (8)
- Basepair signature
- cWW-F-cWW-F-cWW-F-cWW-F
- Heat map statistics
- Min 0.05 | Avg 0.13 | Max 0.29
Coloring options: