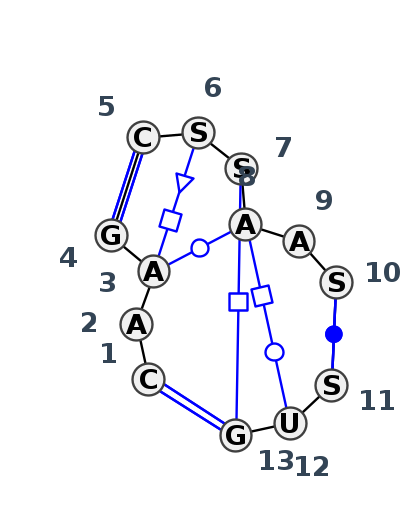
Motif J3_47341.1 Version J3_47341.1 of this group appears in releases 4.0 to 4.7
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | break | 5 | 6 | 7 | 8 | 9 | 10 | break | 11 | 12 | 13 | 1-13 | 3-6 | 3-8 | 4-5 | 5-8 | 7-13 | 8-12 | 10-11 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | J3_9E6Q_009 | 9E6Q | 0.2785 | 1 | 1 | LSU rRNA | C | 1782 | A | 1783 | A | 1784 | G | 1785 | * | C | 1426 | G | 1427 | G | 1428 | A | 1429 | A | 1431 | G | 1432 | * | C | 1768 | U | 1769 | G | 1770 | cWW | tHS | tWW | cWW | ncSs | tHW | cWW | |
| 2 | J3_8B0X_019 | 8B0X | 0.0000 | 1 | a | LSU rRNA | C | 1308 | A | 1309 | A | 1310 | G | 1311 | * | C | 1607 | C | 1608 | C | 1609 | A | 1610 | A | 1612 | C | 1613 | * | G | 1622 | U | 1623 | G | 1624 | cWW | tWW | cWW | ntHH | tHW | cWW | ||
| 3 | J3_9DFE_011 | 9DFE | 0.1159 | 1 | 1A | LSU rRNA | C | 1306 | A | 1307 | A | 1308 | G | 1309 | * | C | 1605 | G | 1606 | C | 1607 | A | 1608 | A | 1610 | C | 1611 | * | G | 1620 | U | 1621 | G | 1622 | cWW | tHS | tWW | cWW | tHH | tHW | cWW | |
| 4 | J3_7A0S_007 | 7A0S | 0.1243 | 1 | X | LSU rRNA | C | 1319 | A | 1320 | A | 1321 | G | 1322 | * | C | 1621 | G | 1622 | C | 1623 | A | 1624 | A | 1626 | C | 1627 | * | G | 1636 | U | 1637 | G | 1638 | cWW | tHS | ntWW | cWW | ntHH | tHW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CAAG*CGCAAAC*GUG | 2 |
| CGGAAAG*CUG*CAAG | 1 |
| CAAG*CCCAAAC*GUG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| AA*GCAAA*U | 2 |
| GGAAA*U*AA | 1 |
| AA*CCAAA*U | 1 |
Release history
| Release | 4.0 | 4.1 | 4.2 | 4.3 | 4.4 | 4.5 | 4.6 | 4.7 |
|---|---|---|---|---|---|---|---|---|
| Date | 2025-08-13 | 2025-09-10 | 2025-10-08 | 2025-11-05 | 2025-12-03 | 2025-12-31 | 2026-01-28 | 2026-02-25 |
| Status | New id, 1 parent | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (4)
- Basepair signature
- cWW-tHH-F-tHW-tHS-tWW-cWW-cWW-F
- Heat map statistics
- Min 0.11 | Avg 0.16 | Max 0.32
Coloring options: