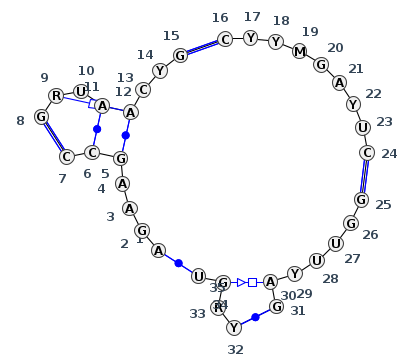
Motif J5_53110.1 Version J5_53110.1 of this group appears in releases 4.0 to 4.6
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | break | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | break | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | break | 25 | 26 | 27 | 28 | 29 | 30 | 31 | break | 32 | 33 | 34 | 35 | 1-35 | 5-12 | 6-11 | 6-19 | 7-8 | 8-10 | 9-12 | 11-19 | 13-18 | 14-18 | 14-22 | 15-16 | 18-22 | 23-28 | 24-25 | 30-34 | 31-32 | |||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | J5_8GLP_004 | 8GLP | 0.1585 | 5 | L5 | LSU rRNA | A | 3903 | G | 3904 | A | 3905 | A | 3906 | G | 3907 | C | 3909 | C | 3910 | * | OMG | 4392 | G | 4393 | U | 4395 | A | 4396 | A | 4397 | C | 4398 | U | 4399 | G | 4400 | * | C | 4444 | U | 4445 | U | 4446 | 5MC | 4447 | G | 4448 | A | 4449 | U | 4450 | U | 4452 | C | 4453 | * | G | 4528 | G | 4529 | UR3 | 4530 | PSU | 4531 | PSU | 4532 | A | 4533 | G | 4534 | * | PSU | 4552 | A | 4553 | G | 4554 | U | 4557 | cWW | ncWW | cWW | ntWS | cWW | ntSW | tHH | tHW | ncWw | ncwW | cWW | cWW | tHS | cWW | |||
| 2 | J5_8OI5_005 | 8OI5 | 0.0000 | 5 | 1 | LSU rRNA | A | 2377 | G | 2378 | A | 2379 | A | 2380 | G | 2381 | C | 2383 | C | 2384 | * | G | 2787 | G | 2788 | U | 2790 | A | 2791 | A | 2792 | C | 2793 | U | 2794 | G | 2795 | * | C | 2839 | U | 2840 | U | 2841 | C | 2842 | G | 2843 | A | 2844 | U | 2845 | U | 2847 | C | 2848 | * | G | 2923 | G | 2924 | U | 2925 | U | 2926 | U | 2927 | A | 2928 | G | 2929 | * | U | 2947 | A | 2948 | G | 2949 | U | 2952 | cWW | ncWW | cWW | ntWS | cWW | ntSW | ntHH | ntHW | cWW | ncSW | ntWWa | cWW | tHS | cWW | |||
| 3 | J5_9E6Q_006 | 9E6Q | 0.1126 | 5 | 1 | LSU rRNA | A | 2177 | G | 2178 | A | 2179 | A | 2180 | G | 2181 | C | 2183 | C | 2184 | * | G | 2560 | A | 2561 | U | 2563 | A | 2564 | A | 2565 | C | 2566 | C | 2567 | G | 2568 | * | C | 2612 | C | 2613 | C | 2614 | A | 2615 | G | 2616 | A | 2617 | C | 2618 | U | 2620 | C | 2621 | * | G | 2696 | G | 2697 | UR3 | 2698 | U | 2699 | C | 2700 | A | 2701 | G | 2702 | * | OMC | 2720 | G | 2721 | G | 2722 | U | 2725 | cWW | cWW | cWW | cWW | ntSW | tHH | ncBW | cWW | cSH | ntSW | cWW | tHS | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| AGAAGACC*OMGGAUAACUG*CUU(5MC)GAUGUC*GG(UR3)(PSU)(PSU)AG*PSUAGUUU | 1 |
| AGAAGACC*GGAUAACUG*CUUCGAUGUC*GGUUUAG*UAGUUU | 1 |
| AGAAGUCC*GAAUAACCG*CCCAGACGUC*GG(UR3)UCAG*OMCGGUCU | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GAAGAC*GAUAACU*UU(5MC)GAUGU*G(UR3)(PSU)(PSU)A*AGUU | 1 |
| GAAGAC*GAUAACU*UUCGAUGU*GUUUA*AGUU | 1 |
| GAAGUC*AAUAACC*CCAGACGU*G(UR3)UCA*GGUC | 1 |
Release history
| Release | 4.0 | 4.1 | 4.2 | 4.3 | 4.4 | 4.5 | 4.6 |
|---|---|---|---|---|---|---|---|
| Date | 2025-08-13 | 2025-09-10 | 2025-10-08 | 2025-11-05 | 2025-12-03 | 2025-12-31 | 2026-01-28 |
| Status | New id, no parents | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (3)
- Basepair signature
- cWW-F-tHS-F-F-F-cWW-cWW-cWW-cWW-F-F-tHH-F-F-F-cWW-F-F-F-F-cWW-F-F-F-F-F
- Heat map statistics
- Min 0.11 | Avg 0.14 | Max 0.37
Coloring options: