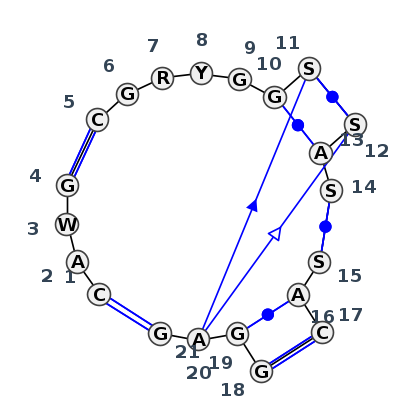
Motif J5_59910.1 Version J5_59910.1 of this group appears in releases 3.2 to 3.17
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | break | 5 | 6 | 7 | 8 | 9 | 10 | 11 | break | 12 | 13 | 14 | break | 15 | 16 | 17 | break | 18 | 19 | 20 | 21 | 1-21 | 3-6 | 4-5 | 10-13 | 11-12 | 11-20 | 12-20 | 14-15 | 16-19 | 17-18 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | J5_5J7L_003 | 5J7L | 0.0998 | 2 | AA | SSU rRNA | C | 58 | A | 59 | A | 60 | G | 61 | * | C | 106 | G | 107 | G | 108 | C | 110 | G | 111 | G | 112 | G | 113 | * | C | 314 | A | 315 | C | 316 | * | G | 337 | A | 338 | C | 339 | * | G | 350 | G | 351 | A | 353 | G | 354 | cWW | ntSs | cWW | cWW | cWW | csS | ntsS | cWW | cWW | cWW |
| 2 | J5_4LFB_002 | 4LFB | 0.0000 | 2 | A | SSU rRNA | C | 58 | A | 59 | A | 60 | G | 61 | * | C | 106 | G | 107 | G | 108 | C | 110 | G | 111 | G | 112 | G | 113 | * | C | 314 | A | 315 | G | 316 | * | C | 337 | A | 338 | C | 339 | * | G | 350 | G | 351 | A | 353 | G | 354 | cWW | ntSs | cWW | cWW | cWW | csS | ntsS | cWW | cWW | cWW |
| 3 | J5_4V88_019 | 4V88 | 0.1520 | 2 | A6 | SSU rRNA | C | 54 | A | 55 | U | 56 | G | 57 | * | C | 90 | G | 91 | A | 92 | U | 94 | G | 95 | G | 96 | C | 97 | * | G | 386 | A | 387 | G | 388 | * | C | 409 | A | 410 | C | 411 | * | G | 422 | G | 423 | A | 425 | G | 426 | cWW | ntWS | cWW | cWW | cWW | csS | tsS | cWW | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CAAG*CGGACGGG*CAC*GAC*GGCAG | 1 |
| CAAG*CGGACGGG*CAG*CAC*GGCAG | 1 |
| CAUG*CGAAUGGC*GAG*CAC*GGCAG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| AA*GGACGG*A*A*GCA | 2 |
| AU*GAAUGG*A*A*GCA | 1 |
Release history
| Release | 3.2 | 3.3 | 3.4 | 3.5 | 3.6 | 3.7 | 3.8 | 3.9 | 3.10 | 3.11 | 3.12 | 3.13 | 3.14 | 3.15 | 3.16 | 3.17 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Date | 2018-02-09 | 2018-03-08 | 2018-04-06 | 2018-05-04 | 2018-06-01 | 2018-06-29 | 2018-07-27 | 2018-08-24 | 2018-09-21 | 2018-10-19 | 2018-11-16 | 2018-12-14 | 2019-01-11 | 2019-02-08 | 2019-03-08 | 2019-04-05 |
| Status | New id, no parents | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (3)
- Basepair signature
- cWW-F-F-F-F-F-cWW-F-cWW-tSS-cSS-cWW-cWW-cWW-cWW
- Heat map statistics
- Min 0.10 | Avg 0.09 | Max 0.17
Coloring options: