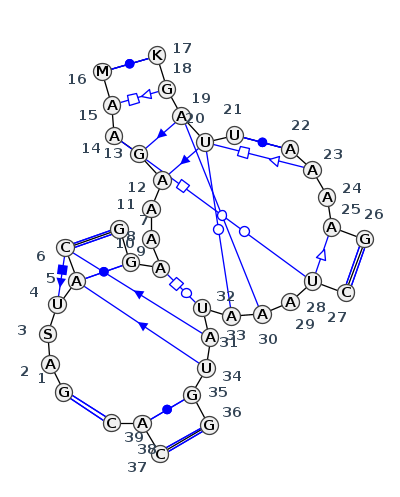
Motif J6_18219.1 Version J6_18219.1 of this group appears in releases 4.0 to 4.0
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | break | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | break | 17 | 18 | 19 | 20 | 21 | break | 22 | 23 | 24 | 25 | 26 | break | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 35 | 36 | break | 37 | 38 | 39 | 1-39 | 4-6 | 5-8 | 5-34 | 6-7 | 6-33 | 7-9 | 9-32 | 12-20 | 13-19 | 14-28 | 15-18 | 16-17 | 19-30 | 20-23 | 20-31 | 21-22 | 23-30 | 25-28 | 26-27 | 35-38 | 36-37 | |||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | J6_9AXU_001 | 9AXU | 0.0782 | 3 | 2+4 | LSU rRNA + 5.8S rRNA | G | 22 | A | 23 | C | 24 | U | 25 | A | 26 | C | 27 | * | G | 58 | G | 59 | A | 60 | A | 61 | A | 62 | A | 63 | G | 64 | A | 65 | A | 66 | A | 68 | * | U | 75 | G | 76 | A | 77 | U | 78 | U | 79 | * | A | 106 | A | 107 | A | 108 | A | 109 | G | 110 | * | C | 329 | U | 330 | A | 331 | A | 332 | A | 333 | U | 334 | A | 335 | U | 336 | G | 338 | G | 339 | * | C | 40 | A | 41 | C | 43 | cWW | cSH | cWW | csS | cWW | cSs | ncsS | tHW | csS | cSs | tHW | tHS | cWW | tWW | tHS | tWW | cWW | tSs | cWW | cWW | cWW | |
| 2 | J6_8P9A_003 | 8P9A | 0.0000 | 3 | AR+AT | LSU rRNA + 5.8S rRNA | G | 22 | A | 23 | G | 24 | U | 25 | A | 26 | C | 27 | * | G | 58 | G | 59 | A | 60 | A | 61 | A | 62 | A | 63 | G | 64 | A | 65 | A | 66 | C | 68 | * | G | 75 | G | 76 | A | 77 | U | 78 | U | 79 | * | A | 106 | A | 107 | A | 108 | A | 109 | G | 110 | * | C | 321 | U | 322 | A | 323 | A | 324 | A | 325 | U | 326 | A | 327 | U | 328 | G | 330 | G | 331 | * | C | 32 | A | 33 | C | 35 | cWW | cWW | csS | cWW | cSs | ncsS | tHW | csS | cSs | tHW | tHS | cWW | tWW | ntHS | tWW | cWW | ncSs | tSs | cWW | cWW | cWW | |
| 3 | J6_8OI5_001 | 8OI5 | 0.0884 | 3 | 1+4 | LSU rRNA + 5.8S rRNA | G | 21 | A | 22 | C | 23 | U | 24 | A | 25 | C | 26 | * | G | 57 | G | 58 | A | 59 | A | 60 | A | 61 | A | 62 | G | 63 | A | 64 | A | 65 | C | 67 | * | G | 74 | G | 75 | A | 76 | U | 77 | U | 78 | * | A | 105 | A | 106 | A | 107 | A | 108 | G | 109 | * | C | 321 | U | 322 | A | 323 | A | 324 | A | 325 | U | 326 | A | 327 | U | 328 | G | 330 | G | 331 | * | C | 32 | A | 33 | C | 35 | cWW | cSH | cWW | csS | cWW | cSs | ncsS | tHW | csS | cSs | tHW | tHS | cWW | tWW | tHS | tWW | cWW | tSs | cWW | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GACUAC*GGAAAAGAAAA*UGAUU*AAAAG*CUAAAUAUUGGCAUC | 1 |
| GAGUAC*GGAAAAGAAAC*GGAUU*AAAAG*CUAAAUAUUGGCAUC | 1 |
| GACUAC*GGAAAAGAAAC*GGAUU*AAAAG*CUAAAUAUUGGCAUC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| ACUA*GAAAAGAAA*GAU*AAA*UAAAUAUUGAU | 2 |
| AGUA*GAAAAGAAA*GAU*AAA*UAAAUAUUGAU | 1 |
Release history
| Release | 4.0 |
|---|---|
| Date | 2025-08-13 |
| Status | New id, 1 parent |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (3)
- Basepair signature
- cWW-F-cWW-F-cWW-cSH-cWW-cSS-cWW-cSS-tHW-tWW-F-tWW-F-F-cSS-tSS-tHW-cSS-cWW-tHS-cWW-F-tHS-cWW
- Heat map statistics
- Min 0.08 | Avg 0.06 | Max 0.09
Coloring options: