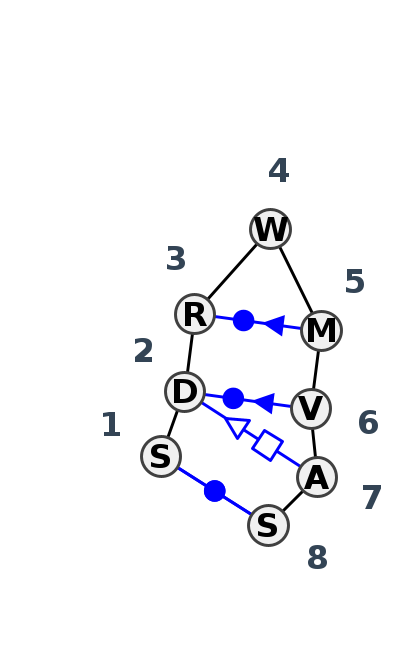
Motif HL_18470.2 Version HL_18470.2 of this group appears in releases 4.1 to 4.6
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 1-8 | 2-6 | 2-7 | 3-5 | 3-6 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HL_8OI5_014 | 8OI5 | 0.9572 | 1 | 1 | LSU rRNA | G | 684 | U | 685 | G | 686 | A | 688 | A | 689 | A | 690 | A | 691 | C | 692 | cWW | ncSW | tSH | |||
| 2 | HL_8P9A_127 | 8P9A | 0.9433 | 1 | AR | LSU rRNA | G | 686 | U | 687 | G | 688 | A | 690 | A | 691 | A | 692 | A | 693 | C | 694 | cWW | ncSW | tSH | |||
| 3 | HL_3IGI_010 | 3IGI | 0.8971 | 2 | A | Group II catalytic intron D1-D4-3 | C | 305 | U | 306 | G | 307 | U | 308 | C | 310 | C | 312 | A | 313 | G | 314 | ncWW | ncWS | ||||
| 4 | HL_2BTE_005 | 2BTE | 0.1328 | 5 | tRNA D-loop (H) | E | tRNA | G | 12 | G | 13 | A | 14 | A | 15 | A | 20|||A | G | 21 | A | 22 | C | 23 | cWW | cWS | tSH | cWS | |
| 5 | HL_9J5P_001 | 9J5P | 0.0000 | 5 | A | tRNA | G | 12 | G | 13 | A | 14 | A | 15 | A | 21 | G | 22 | A | 23 | C | 24 | cWW | cWS | tSH | cWS | ||
| 6 | HL_3ZGZ_001 | 3ZGZ | 0.1010 | 5 | tRNA D-loop | B | tRNA | G | 12 | G | 13 | A | 14 | A | 15 | A | 20|||A | G | 21 | A | 22 | C | 23 | cWW | cWS | tSH | cWS | |
| 7 | HL_5AH5_005 | 5AH5 | 0.1294 | 5 | tRNA D-loop | D | tRNA | G | 12 | A | 13 | A | 14 | A | 15 | A | 20|||A | A | 21 | A | 22 | C | 23 | cWW | cWS | tSH | cWS |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GUGUAAAAC | 2 |
| CUGUUCGCAG | 1 |
| GGAAUGGGUAGAC | 1 |
| GGAAUUGGCAGAC | 1 |
| GGAAUCGGUAGAC | 1 |
| GAAAUCGGUAAAC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| UGUAAAA | 2 |
| UGUUCGCA | 1 |
| GAAUGGGUAGA | 1 |
| GAAUUGGCAGA | 1 |
| GAAUCGGUAGA | 1 |
| AAAUCGGUAAA | 1 |
- Annotations
-
- (4)
- tRNA D-loop (2)
- tRNA D-loop (H) (1)
- Basepair signature
- cWW-cWS-tSH-cWS-F
- Heat map statistics
- Min 0.06 | Avg 0.55 | Max 0.98
Coloring options: