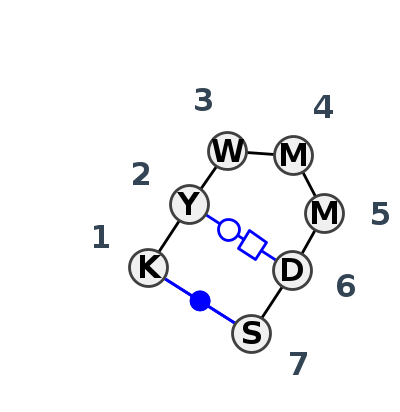
Motif HL_20554.2 Version HL_20554.2 of this group appears in releases 3.90 to 3.95
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 1-7 | 2-6 | 5-6 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HL_5J7L_153 | 5J7L | 0.9289 | 0 | DA | LSU rRNA | U | 641 | U | 642 | A | 643 | A | 644 | C | 645 | U | 646 | G | 647 | cWW | ncSH | ||
| 2 | HL_2A43_001 | 2A43 | 0.2467 | 1 | A | Putative RNA-dependent RNA polymerase ribosomal frameshift site | G | 6 | C | 7 | A | 8 | C | 9 | C | 10 | G | 11 | C | 13 | cWW | tWH | ||
| 3 | HL_5J7L_014 | 5J7L | 0.1304 | 0 | GNRA-like with tWH | AA | SSU rRNA | G | 617 | C | 618 | U | 619 | C | 620 | A | 621 | A | 622 | C | 623 | cWW | ntWH | |
| 4 | HL_4LFB_014 | 4LFB | 0.0000 | 0 | GNRA-like with tWH | A | SSU rRNA | G | 617 | C | 618 | U | 619 | C | 620 | A | 621 | A | 622 | C | 623 | cWW | tWH | |
| 5 | HL_6CZR_085 | 6CZR | 0.2023 | 0 | 1a | SSU rRNA | G | 601 | C | 602 | U | 603 | C | 604 | A | 605 | A | 606 | C | 607 | cWW | ntWH |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GCUCAAC | 3 |
| UUAACUG | 1 |
| GCACCGUC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| CUCAA | 3 |
| UAACU | 1 |
| CACCGU | 1 |
- Annotations
-
- (3)
- GNRA-like with tWH (2)
- Basepair signature
- cWW-tWH-F-F-F
- Heat map statistics
- Min 0.13 | Avg 0.42 | Max 0.97
Coloring options: