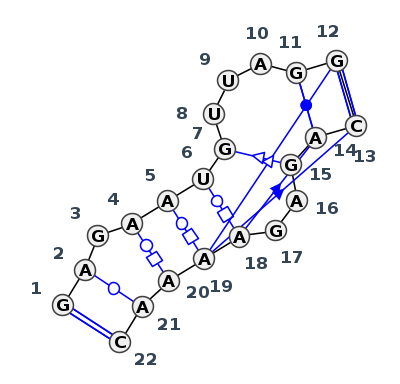
Motif IL_58354.1 Version IL_58354.1 of this group appears in releases 4.1 to 4.6
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | break | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 1-22 | 2-21 | 2-22 | 4-20 | 5-19 | 6-18 | 7-15 | 11-14 | 11-15 | 12-13 | 12-19 | 13-19 | 14-18 | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_6SY6_001 | 6SY6 | 0.0000 | 1 | Kink-turn related | D | RNA (36-MER) | G | 5 | A | 6 | G | 7 | A | 8 | A | 9 | U | 10 | G | 11 | U | 12 | U | 13 | A | 14 | G | 16 | G | 17 | * | C | 26 | A | 27 | G | 28 | A | 29 | G | 30 | A | 31 | A | 32 | A | 33 | A | 34 | C | 35 | cWW | ntWW | ntWW | tWH | tWH | tWH | tSS | ncWW | ntSH | cWW | tsS | csS | ncSs |
| 2 | IL_6SY4_001 | 6SY4 | 0.0935 | 1 | Kink-turn related | C | TetR-binding aptamer K1 (43-MER) | G | 6 | A | 7 | G | 8 | A | 9 | A | 10 | U | 11 | G | 12 | U | 13 | U | 14 | A | 15 | G | 17 | G | 18 | * | C | 29 | A | 30 | G | 31 | A | 32 | G | 33 | A | 34 | A | 35 | A | 36 | A | 37 | C | 38 | cWW | tWW | tWH | tWH | tWH | tSS | cWW | ntSH | cWW | tSs | csS | cSs |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GAGAAUGUUAUGG*CAGAGAAAAC | 2 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| AGAAUGUUAUG*AGAGAAAA | 2 |
- Annotations
-
- Kink-turn related (2)
- Basepair signature
- cWW-tWW-L-tWH-cSS-tSS-tWH-cSS-tWH-R-tSS-R-L-L-cWW-L-cWW
- Heat map statistics
- Min 0.09 | Avg 0.05 | Max 0.09
Coloring options: