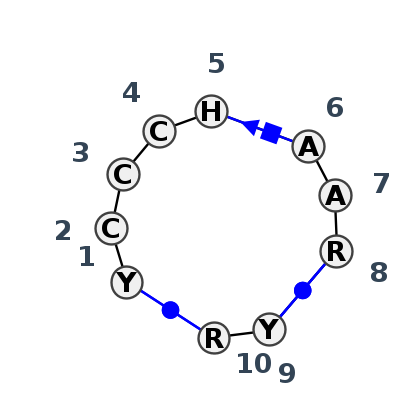
Motif IL_71457.1 Version IL_71457.1 of this group appears in releases 3.88 to 3.88
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | break | 9 | 10 | 1-10 | 5-6 | 8-9 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_7A0S_034 | 7A0S | 0.1785 | 0 | SSU/LSU pseudoknot | X | LSU rRNA | U | 1015 | C | 1016 | C | 1017 | C | 1018 | U | 1019 | A | 1020 | A | 1021 | A | 1022 | * | U | 1161 | A | 1162 | cWW | ncSH | cWW |
| 2 | IL_5J7L_278 | 5J7L | 0.1237 | 0 | SSU/LSU pseudoknot | DA | LSU rRNA | U | 1004 | C | 1005 | C | 1006 | C | 1007 | A | 1008 | A | 1009 | A | 1010 | G | 1011 | * | C | 1150 | A | 1151 | cWW | cSH | cWW |
| 3 | IL_4WF9_037 | 4WF9 | 0.1522 | 0 | SSU/LSU pseudoknot | X | LSU rRNA | U | 1048 | C | 1049 | C | 1050 | C | 1051 | A | 1052 | A | 1053 | A | 1054 | A | 1055 | * | U | 1194 | A | 1195 | cWW | ncSH | cWW |
| 4 | IL_8VTW_036 | 8VTW | 0.0000 | 0 | SSU/LSU pseudoknot | 1A | LSU rRNA | C | 1004 | C | 1005 | C | 1006 | C | 1007 | C | 1008 | A | 1009 | A | 1010 | G | 1011 | * | C | 1150 | G | 1151 | cWW | cSH | cWW |
| 5 | IL_4V9F_038 | 4V9F | 0.1214 | 0 | SSU/LSU pseudoknot | 0 | LSU rRNA | U | 1101 | C | 1102 | C | 1103 | C | 1104 | C | 1105 | A | 1106 | A | 1107 | G | 1108 | * | C | 1254 | A | 1255 | cWW | cSH | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| UCCCUAAA*UA | 1 |
| UCCCAAAG*CA | 1 |
| UCCCAAAA*UA | 1 |
| CCCCCAAG*CG | 1 |
| UCCCCAAG*CA | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| CCCAAA* | 2 |
| CCCCAA* | 2 |
| CCCUAA* | 1 |
Release history
| Release | 3.88 |
|---|---|
| Date | 2024-09-11 |
| Status | New id, 1 parent |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- SSU/LSU pseudoknot (5)
- Basepair signature
- cWW-L-cWW-L-L-R-cSH
- Heat map statistics
- Min 0.09 | Avg 0.14 | Max 0.26
Coloring options: