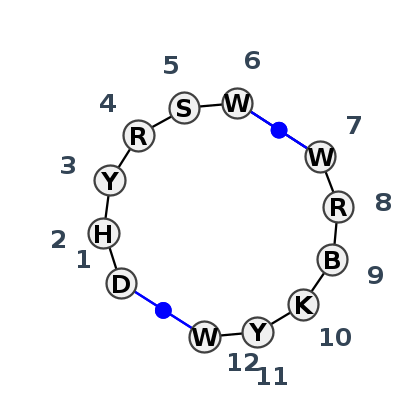
Motif IL_95349.1 Version IL_95349.1 of this group appears in releases 4.0 to 4.6
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | break | 7 | 8 | 9 | 10 | 11 | 12 | 1-12 | 2-11 | 3-10 | 4-9 | 5-8 | 6-7 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_3IGI_015 | 3IGI | 0.4018 | 0 | A | Group II catalytic intron D1-D4-3 | U | 214 | A | 215 | C | 216 | A | 217 | G | 218 | A | 219 | * | U | 158 | A | 159 | G | 160 | G | 161 | U | 162 | A | 163 | ncWW | ncWW | ||||
| 2 | IL_9AXU_040 | 9AXU | 0.0000 | 0 | 2 | LSU rRNA | A | 1038 | U | 1039 | U | 1040 | A | 1041 | G | 1042 | A | 1043 | * | U | 1071 | A | 1072 | U | 1073 | U | 1074 | C | 1075 | U | 1076 | ncWW | ncWw | ncWW | cWW | cWW | |
| 3 | IL_9AXU_008 | 9AXU | 0.4291 | 0 | 2 | LSU rRNA | G | 167 | C | 168 | U | 169 | G | 170 | C | 171 | U | 172 | * | A | 263 | G | 264 | C | 265 | G | 266 | C | 267 | U | 268 | ncWWa | cWw | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| UACAGA*UAGGUA | 1 |
| AUUAGA*UAUUCU | 1 |
| GCUGCU*AGCGCU | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| ACAG*AGGU | 1 |
| UUAG*AUUC | 1 |
| CUGC*GCGC | 1 |
Release history
| Release | 4.0 | 4.1 | 4.2 | 4.3 | 4.4 | 4.5 | 4.6 |
|---|---|---|---|---|---|---|---|
| Date | 2025-08-13 | 2025-09-10 | 2025-10-08 | 2025-11-05 | 2025-12-03 | 2025-12-31 | 2026-01-28 |
| Status | New id, 1 parent | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (3)
- Basepair signature
- cWW-L-R-L-R-L-R-L-R-cWW
- Heat map statistics
- Min 0.40 | Avg 0.32 | Max 0.61
Coloring options: