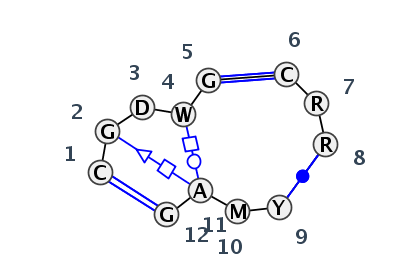
Motif J3_32852.3 Version J3_32852.3 of this group appears in releases 3.54 to 3.86
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | break | 6 | 7 | 8 | break | 9 | 10 | 11 | 12 | 1-12 | 2-11 | 4-7 | 4-10 | 4-11 | 5-6 | 8-9 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | J3_5J7L_041 | 5J7L | 0.2035 | 1 | DA | LSU rRNA | C | 1298 | G | 1299 | G | 1300 | A | 1301 | G | 1303 | * | C | 1625 | A | 1626 | G | 1627 | * | C | 1639 | A | 1640 | A | 1641 | G | 1642 | cWW | tSH | ntHW | cWW | cWW | ||
| 2 | J3_4WF9_004 | 4WF9 | 0.1543 | 1 | X | LSU rRNA | C | 1335 | G | 1336 | A | 1337 | U | 1338 | G | 1340 | * | C | 1669 | A | 1670 | A | 1671 | * | U | 1683 | A | 1684 | A | 1685 | G | 1686 | cWW | tSH | ntHH | tWW | cWW | cWW | |
| 3 | J3_7RQB_007 | 7RQB | 0.0000 | 1 | 1A | LSU rRNA | C | 1298 | G | 1299 | U | 1300 | A | 1301 | G | 1303 | * | C | 1625 | G | 1626 | G | 1627 | * | U | 1639 | C | 1640 | A | 1641 | G | 1642 | cWW | tSH | tSs | ntHW | cWW | cWW | |
| 4 | J3_7A0S_006 | 7A0S | 0.0880 | 1 | X | LSU rRNA | C | 1311 | G | 1312 | U | 1313 | A | 1314 | G | 1316 | * | C | 1641 | G | 1642 | A | 1643 | * | U | 1656 | A | 1657 | A | 1658 | G | 1659 | cWW | tSH | tSs | ntHW | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CGGAAG*CAG*CAAG | 1 |
| CGAUUG*CAA*UAAG | 1 |
| CGUAAG*CGG*UCAG | 1 |
| CGUAAG*CGA*UAAG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GGAA*A*AA | 1 |
| GAUU*A*AA | 1 |
| GUAA*G*CA | 1 |
| GUAA*G*AA | 1 |
Release history
| Release | 3.54 | 3.55 | 3.56 | 3.57 | 3.58 | 3.59 | 3.60 | 3.61 | 3.62 | 3.63 | 3.64 | 3.65 | 3.66 | 3.67 | 3.68 | 3.69 | 3.70 | 3.71 | 3.72 | 3.73 | 3.74 | 3.75 | 3.76 | 3.77 | 3.78 | 3.79 | 3.80 | 3.81 | 3.82 | 3.83 | 3.84 | 3.85 | 3.86 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Date | 2022-02-02 | 2022-03-02 | 2022-03-30 | 2022-04-27 | 2022-05-25 | 2022-06-22 | 2022-07-20 | 2022-08-17 | 2022-10-12 | 2022-11-09 | 2022-12-07 | 2023-01-04 | 2023-02-01 | 2023-03-01 | 2023-03-29 | 2023-04-26 | 2023-05-24 | 2023-06-21 | 2023-07-19 | 2023-08-16 | 2023-09-13 | 2023-10-11 | 2023-11-08 | 2023-12-06 | 2024-01-03 | 2024-01-03 | 2024-01-31 | 2024-02-28 | 2024-03-27 | 2024-04-24 | 2024-05-22 | 2024-06-19 | 2024-07-17 |
| Status | Updated, 1 parent | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (4)
- Basepair signature
- cWW-tSH-tHW-F-F-cWW-cWW-F
- Heat map statistics
- Min 0.09 | Avg 0.13 | Max 0.22
Coloring options: