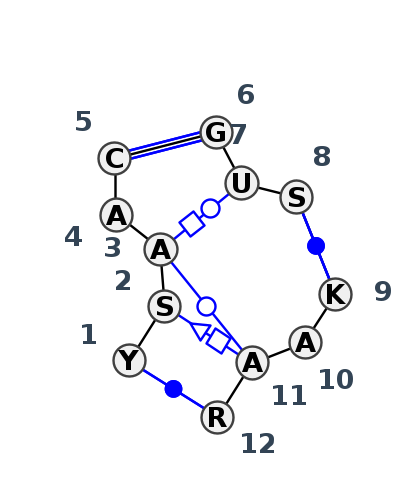
Motif J3_74051.1 Version J3_74051.1 of this group appears in releases 3.89 to 3.90
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | break | 6 | 7 | 8 | break | 9 | 10 | 11 | 12 | 1-12 | 2-11 | 3-7 | 3-11 | 5-6 | 8-9 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | J3_8C3A_007 | 8C3A | 0.0956 | 2 | 1 | LSU rRNA | U | 1833 | C | 1834 | A | 1835 | A | 1838 | C | 1839 | * | G | 1848 | U | 1849 | C | 1850 | * | G | 1484 | A | 1485 | A | 1486 | A | 1487 | cWW | ntSH | tHW | ntWW | cWW | cWW |
| 2 | J3_5TBW_009 | 5TBW | 0.0000 | 2 | 1 | LSU rRNA | U | 1837 | G | 1838 | A | 1839 | A | 1842 | C | 1843 | * | G | 1852 | U | 1853 | C | 1854 | * | G | 1488 | A | 1489 | A | 1490 | A | 1491 | cWW | ntSH | tHW | ntWW | cWW | cWW |
| 3 | J3_4V9F_006 | 4V9F | 0.1050 | 2 | 0 | LSU rRNA | C | 1680 | G | 1681 | A | 1682 | A | 1685 | C | 1686 | * | G | 1695 | U | 1696 | G | 1697 | * | U | 1412 | A | 1413 | A | 1414 | G | 1415 | cWW | tSH | tHW | tWW | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GAAA*UCAUAAC*GUC | 1 |
| GAAA*UGAUAAC*GUC | 1 |
| UAAG*CGAGAAC*GUG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| AA*CAUAA*U | 1 |
| AA*GAUAA*U | 1 |
| AA*GAGAA*U | 1 |
- Annotations
-
- (3)
- Basepair signature
- cWW-tSH-tWW-tHW-F-F-cWW-cWW
- Heat map statistics
- Min 0.10 | Avg 0.07 | Max 0.12
Coloring options: