JAR3D
Query a223a5a5-67f9-4011-be6c-60be02d50134 completed
Run on Motif Atlas Version 1.18
Mitochondria, SSU, helix 27, mammals only
Input Summary
0000000001111111111222222222
1234567890123456789012345678
(((------((((----))))----)))
>260331.1 Sus scrofa
GAGUACUACUCGCAACUGCCUAAAACUC
>174520.1 Nasalis larvatus
GAACACUGCAAGCAACAGCUUAAAACUC
>55470.1 Cavia porcellus
GAGAACUACUAGCAAUAGCUUAAAACUC
>253360.1 Stenella coeruleoalba
GAGUACUAUCGGCAACAGCCA-AAACUC
>129339.1 Homo sapiens
GAACACUACGAGCCACAGCUUAAAACUC
>260335.1 Sus scrofa
GAGUACUACUCGCAACUGCCUAAAACUC
>260358.1 Sus scrofa
GAGUACUACUCGCAACUGCCUAAAACUC
>357.1 Abrothrix jelskii
GAGAACUACUGGCCACAGCUUAAAACUC
>2159.1 Acerodon celebensis
GAGUACUACUAGCAACAGCUUAAAACUC
>3771.1 Acomys cahirinus
GAGAACUACUAGCCAUAGCUUAAAACUC
>6925.1 Aepyceros melampus
GAGUACUACCGGCAACGGCCCAAAACUC
>6927.1 Aepyprymnus rufescens
GAGAACUACAAGCCAAUGCUUAAAACUC
>9610.1 Ailuropoda melanoleuca
GAGAACUACUAGCAACAGCUUAAAACUC
>9617.1 Ailurus fulgens
GAGAACUACUAGCAAUAGCCUAAAACUC
>11824.1 Alouatta palliata
GAACACUACAAGCAACAGCUUGAAACUC
>11825.1 Alouatta seniculus
GAACACUACAAGCAACAGCUUGAAAUUC
>13475.1 Amblysomus hottentotus
GAGAACUACUAGCAAUAGCCUAAAACUU
>18456.1 Antechinomys laniger
GAGAACUACGAGCCACCGCUUAAAACUC
>18457.1 Antechinus bellus
GAGAACUACAAGCCACUGCUUAAAACUC
>18458.1 Antechinus flavipes
GAGAACUACAAGCCACUGCUUAAAACUC
>18459.1 Antechinus godmani
GAGAACUACAAGCCACUGCUUAAAACUC
>18460.1 Micromurexia habbema
GAGAACUACAAGCCACUGCUUAAAACUC
>18461.1 Antechinus leo
GAGAACUACAAGCCACUGCUUAAAACUC
>18462.1 Murexchinus melanurus
GAGAACUACAAGCCACUGCUUAAAACUC
>18463.1 Antechinus minimus
GAGAACUACAAGCCACUGCUUAAAACUC
>18464.1 Antechinus naso
GAGAACUACAAGCUACUGCUUAAAACUC
>18465.1 Antechinus sp.
GAGAACUACAAGCCACUGCUUAAAACUC
>18466.1 Antechinus stuartii
GAGAACUACAAGCUACUGCUUAAAACUC
>18467.1 Antechinus stuartii
GAGAACUACAAGCCACUGCUUAAAACUC
>18468.1 Antechinus swainsonii
GAGAACUACGAGCCACUGCUUAAAACUC
>18469.1 Antechinus swainsonii
GAGAACUACGAGCCACUGCUUAAAACUC
>18717.1 Antilocapra americana
GAGUACUACUAGCAACUGCUUAAAACUC
>18984.1 Aotus trivirgatus
GAAUACUACAAGCCACAGCUUGAAACUC
>20242.1 Aproteles bulmerae
GAGUACUACGAGCAACAGCUUAAAACUC
>21866.1 Arctocephalus forsteri
GAGAACUACUAGCAACAGCUUAAAACUC
>21867.1 Arctocephalus gazella
GAGAACUACUAGCAACAGCUUAAAACUC
>24636.1 Artibeus jamaicensis
GAGUACUACUAGCCAAAGCUUAAAACUC
>26910.1 Atelerix albiventris
GAGAACUACGAGCCACAGCUUAAAACUC
>26914.1 Ateles sp.
GAACACUACAAGCAACAGCUUGAAACUC
>34517.1 Balaenoptera musculus
GAGUACUACUAGCAACAGCUUAAAACUC
>34522.1 Balaenoptera physalus
GAGUACUACUAGCAACAGCCUAAAACUC
>35156.1 Bassaricyon gabbii
GAGAACUACUAGCAACAGCUUAAAACUC
>36857.1 Bettongia penicillata
GAGAACUACUAGCCAAUGCUUAAAACUC
>38198.1 Blarina brevicauda
GAGGACUACUAGCAACAGCUUAAAACUC
>40459.1 Bos taurus
GAGUACUACUAGCAACAGCUUAAAACUC
>40640.1 Boselaphus tragocamelus
GAGUACUACCGGCAACAGCUUAAAACUC
>42382.1 Brachyteles arachnoides
GAACACUACAAGCAACAGCUUGAAACUC
>42640.1 Bradypus tridactylus
GAGUACUACUAGCAAUAGCUUGAAACUC
>47400.1 Burramys parvus
GAGAACUACUAGCCAGCGCUUAAAACUC
>48001.1 Caenolestes fuliginosus
GAGAACUACUAGCAAUCGCUUAAAACUC
>48726.1 Callicebus moloch
GAACACUACAAGCCACAGCUUGAAACUC
>48768.1 Callimico goeldii
GAACACUACAAGCAACAGCUUGAAAUUC
>48891.1 Callithrix jacchus
GAACACUACAAGCAACAGCUUGAAAUUC
>48893.1 Callithrix pygmaea
GAACACUACAAGCAACAGCUUGAAACUC
>49390.1 Caluromys philander
GAGAACUACUAGCAACUGCUUAAAACUC
>49595.1 Camelus bactrianus
GAGUACUACUAGCAACAGCUUAAAACUC
>53209.1 Canis lupus familiaris
GAGGACUACUAGCAAUAGCUUAAAACUC
>53210.1 Canis lupus familiaris
GAGGACUACUAGCAAUAGCUUAAAACUC
>53634.1 Capra hircus
GAGUACUACCGGCAACAGCCCGAAACUC
>55487.1 Cebus apella
GAACACUACAAGCAAUAGCUUAAAACUC
>56070.1 Philantomba maxwellii
GAGUACUACCGGCAACAGCCUAAAACUC
>57005.1 Ceratotherium simum
GAGUACUACUAGCAACAGCCUAAAACUC
>57006.1 Ceratotherium simum
GAGUACUACUAGCAACAGCCUAAAACUC
>57677.1 Rusa unicolor
GAGUACUACCGGCAAUAGCUUAAAACUC
>57994.1 Chaeropus ecaudatus
GAGAACUACUAGCAAAUGCUUGAAACUC
>58280.1 Chaetophractus villosus
GAGUACUACUAGCAAUAGCUUAAAACUC
>59651.1 Chiropotes satanas
GAACACUACAAGCAACAGCUUGAAAUUC
>65299.1 Myodes glareolus
GAGAACUACUAGCCACAGCUUAAAACUC
>74236.1 Cricetomys gambianus
GAGAACUACUAGCCACAGCCUAAAACUC
>74251.1 Cricetulus migratorius
GAGAACUACUAGCCACCGCUUAAAACUC
>79272.1 Galeopterus variegatus
GAGUACUACCAGCAACGGCCCAAAACUC
>79336.1 Cynopterus brachyotis
GAGAACUACUAGCAACAGCUUAAAACUC
>80603.1 Damaliscus pygargus
GAGUACUACAGGCCACAGCCCAAAACUC
>81228.1 Dasycercus cristicauda
GAGAACUACUAGCCACUGCUUAAAACUC
>81269.1 Dasypus novemcinctus
GAGUACUACUAGCAACAGCUUAAAACUC
>81283.1 Dasyuroides byrnei
GAGAACUACUAGCCACUGCUUAAAACUC
>81286.1 Dasyurus albopunctatus
GAGAACUACUAGCCACUGCUUAAAACUC
>81287.1 Dasyurus geoffroii
GAGAACUACUAGCCACUGCUUAAAACUC
>81294.1 Dasyurus maculatus
GAGAACUACUAGCCACUGCUUAAAACUC
>81296.1 Dasyurus spartacus
GAGAACUACUAGCCACUGCUUAAAACUC
>82596.1 Dendrolagus dorianus
GAGAACUACUAGCCAAUGCUUAAAACUC
>82598.1 Dendrolagus goodfellowi
GAGAACUACUAGCCAACGCUUAAAACUC
>84839.1 Didelphis virginiana
GAGAACUACUAGCAAUUGCUUAAAACUC
>87122.1 Dobsonia moluccensis
GAGCACUACUAGCAAUAGCUUAAAACUC
>87389.1 Dorcopsis veterum
GAGAACUACUAGCCAGCGCUUAAAACUC
>87390.1 Dorcopsulus vanheurni
GAGAACUACUAGCCAGCGCUUAAAACUC
>88000.1 Dromiciops gliroides
GGGAACUACUAGCUAAUGCUUAAAACUC
>88656.1 Dryomys nitedula
GAGUACUACAAGCCAUAGCUCAAAACUC
>88721.1 Dugong dugon
GAGAACUACUAGCAAAAGCUUAAAACUC
>89677.1 Echinops telfairi
GAGAACUACUAGCAAA-GCUAAAAACUU
>89768.1 Echymipera kalubu
GAGUACUACUAGCAAAUGCUUAAAACUC
>90131.1 Eidolon dupraenum
GAGUACUACGGGCAACAGCUUAAAACUC
>90709.1 Elephantulus rufescens
GAGAACUACAAGCCACUGCUUAAAACUC
>90716.1 Elephas maximus
GAGAACUACUAGCCAGAGCUUAAAACUU
>90717.1 Elephas maximus
GAGAACUACUAGCCAGAGCUUAAAACUU
>90718.1 Elephas maximus
GAGAACUACUAGCCAGAGCUUAAAACUU
>90917.1 Eliomys quercinus
GAGAACUACGAGCCACAGCUCAAAACUC
>92269.1 Enhydra lutris
GAGAACUACUAGCAAUAGCUUAAAACUC
>93527.1 Eonycteris spelaea
GAGCACUACUAGCAACAGCUUAAAACUC
>94418.1 Epomophorus wahlbergi
GAGCACUACUAGCAACAGCCUAAAACUC
>94448.1 Eptesicus fuscus
GAGUACUACCAGCAAGGGCUAGAAACUC
>94594.1 Equus asinus
GAGUACUACUAGCAACAGCCUAAAACUC
>94616.1 Equus caballus
GAGUACUACUAGCAACAGCCUAAAACUC
>94619.1 Equus grevyi
GAGUACUACUAGCAACAGCCUAAAACUC
>95092.1 Erinaceus europaeus
GAGAACUACGAGCUACAGCUUAAAACUC
>101864.1 Felis catus
GAGAACUACUAGCAAUAGCUUAAAACUC
>101865.1 Felis catus
GAGAACUACUAGCAAUAGCUUAAAACUC
>108185.1 Eudorcas thomsonii
GAGUACUACCGGCAACAGCCUAAAACUC
>109697.1 Gerbillus nigeriae
GAGAACUACUAGCCAACGCUUAAAACUC
>110439.1 Glaucomys volans
GAGUACUACUAGCAAUUGCUUAAAACUC
>115894.1 Gorilla gorilla
GAACACUACGAGCCACAGCUUAAAACUC
>115896.1 Gorilla gorilla
GAACACUACGAGCCACAGCUUAAAACUC
>116395.1 Graphiurus murinus
GAGAACUAUAGGCAACAGC-AAAAACUC
>119154.1 Halichoerus grypus
GAGAACUACUAGCAACAGCUUAAAACUC
>124548.1 Herpestes auropunctatus
GAGAACUACUAGCAACAGCUUAAAACUC
>125413.1 Hexaprotodon liberiensis
GAGUACUACUAGCAAUAGCUUAAAACUC
>125761.1 Hippopotamus amphibius
GAGUACUACUAGCAACAGCUUAAAACUC
>125762.1 Hippopotamus amphibius
GAGUACUACUAGCAACAGCUUAAAACUC
>125803.1 Hipposideros galeritus
GAGUACUACCGGCAACAGCCUAAAACUC
>129340.1 Homo sapiens
GAACACUACGAGCCACAGCUUAAAACUC
>129341.1 Homo sapiens
GAACACUACGAGCCACAGCUUAAAACUC
>129342.1 Homo sapiens
GAACACUACGAGCCACAGCUUAAAACUC
>129343.1 Homo sapiens
GAACACUACGAGCCACAGCUUAAAACUC
>129344.1 Homo sapiens
GAACACUACGAGCCACAGCUUAAAACUC
>132763.1 Hydrochoerus hydrochaeris
GAGAACUACUAGCAACAGCUUAAAACUC
>132958.1 Hydropotes inermis
GAGUACUACCGGCAAUAGCUUAAAACUC
>133663.1 Hylobates lar
GAACACUACGAGCAACAGCUUAAAAAUC
>133664.1 Hylobates lar
GAACACUACGAGCAACAGCUUAAAAAUC
>133706.1 Hylomyscus stella
GAGAACUACUAGCCAUAGCUUAAAACUC
>135416.1 Hypsiprymnodon moschatus
GAGAACUACUAGCAAUCGCUUAAAACUC
>137245.1 Isoodon macrourus
GAGAACUACUAGCAAAUGCUUAAAACUC
>139772.1 Kobus ellipsiprymnus
GAGUACUACCGGCAAUAGCCUAAAACUC
>143147.1 Lagorchestes hirsutus
GAGAACUACUAGCCAACGCUUAAAACUC
>143152.1 Lagothrix lagotricha
GAACACUACAAGCAAUAGCUUGAAACUC
>143198.1 Lama guanicoe
GAGUACUACUAGCAACAGCUUAAAACUC
>145343.1 Lemur catta
GAGCACUACAAGCAACAGCUUAAAACUC
>145843.1 Leontopithecus rosalia
GAACACUACAAGCAACAGCUUGAAACUC
>145849.1 Leopoldamys edwardsi
GAGAACUACUAGCCACAGCUUAAAACUC
>147417.1 Leptonychotes weddellii
GAGAACUACUAGCAACAGCUUAAAACUC
>151949.1 Loxodonta africana
GAGAACUACUAGCCAGAGCUUAAAACUU
>151950.1 Loxodonta africana
GAGAACUACUAGCCAGAGCUUAAAACUU
>151951.1 Loxodonta africana
GAGAACUACUAGCCAGAGCUUAAAACUU
>152348.1 Lutreolina crassicaudata
GAGGACUACUAGCCAUUGCUUAAAACUC
>154472.1 Macroglossus minimus
GAGAACUACUAGCUACAGCUUAAAACUC
>154809.1 Macropus agilis
GAGAACUACUAGCCAAUGCUUAAAACUC
>154816.1 Macropus giganteus
GAGAACUACUAGCCAAUGCUUAAAACUC
>154822.1 Macropus robustus
GAGAACUACUAGCCAAUGCUUAAAACUC
>154828.1 Macropus rufus
GAGAACUACUAGCCAGUGCUUAAAACUC
>154859.1 Macrotarsomys ingens
GAGAACUACUAGCCACAGCUUAAAACUC
>154870.1 Macrotis lagotis
GAGAACUACUAGCAAAUGCUUGAAACUC
>154942.1 Madoqua kirkii
GAGUACUACCGGCAACAGCCUAAAACUC
>155848.1 Mammuthus primigenius
GAGAACUACUAGCCAGAGCUUAAAACUU
>156075.1 Manis sp.
GAGUACUACUAGCAAUAGCUUGAAACUC
>157872.1 Mastomys erythroleucus
GAGAACUACUAGCCACAGCUUAAAACUC
>160213.1 Megaderma lyra
GAGUACUACGGACAACAGUCUAAAACUC
>160269.1 Megaloglossus woermanni
GAGUACUACUAGCAACAGCUUAAAACUC
>161283.1 Meles meles
GAGAACUACUAGCAACAGCCUAAAACUC
>161738.1 Melonycteris fardoulisi
GAGUACUACUAGCAACAGCUUAAAACUC
>161895.1 Mephitis mephitis
GAGAACUACUAGCAACAGCUUAAAACUC
>162394.1 Mesocricetus auratus
GAGAACUACUGGCCACAGCUUAAAACUC
>166008.1 Microperoryctes longicauda
GAGAACUACUAGCAAAUGCUUAAAACUC
>166464.1 Chionomys nivalis
GAGAACUACUGGCCACAGCUUAAAACUC
>166968.1 Mirounga leonina
GAGAACUACUAGCAACAGCUUAAAACUC
>167513.1 Molossus sinaloae
GAGUACUACUAGCAAAAGCUUAAAACUC
>167555.1 Monachus schauinslandi
GAGAACUACUAGCAAUAGCUUAAAACUC
>168239.1 Monodelphis domestica
GGAAACUACUAGCUACUGCUUAAAACUC
>169684.1 Muntiacus reevesi
GAGUACUACCGGCAAUAGCUUAAAACUC
>169909.1 Murexia longicaudata
GAGAACUACAAGCUACUGCUUAAAACUC
>169911.1 Paramurexia rothschildi
GAGAACUACAAGCUACUGCUUAAAACUC
>169978.1 Mus cookii
GAGAACUACUAGCCAUAGCUUAAAACUC
>169979.1 Mus crociduroides
GAGAACUACUAGCCACAGCUUAAAACUC
>169997.1 Mus mattheyi
GAGAACUACUAGCUACAGCUUAAAACUC
>170039.1 Mus musculus
GAGAACUACUAGCCAUAGCUUAAAACUC
>170040.1 Mus musculus
GAGAACUACUAGCCAUAGCUUAAAACUC
>170041.1 Mus musculus
GAGAACUACUAGCCAUAGCUUAAAACUC
>170042.1 Mus musculus
GAGAACUACUAGCCAUAGCUUAAAACUC
>170043.1 Mus musculus
GAGAACUACUAGCCAUAGCUUAAAACUC
>170078.1 Mus pahari
GAGAACUACUAGCUACAGCUUAAAACUC
>170080.1 Mus platythrix
GAGAACUACUAGCUAUAGCUUAAAACUC
>170081.1 Mus saxicola
GAGAACUACUAGCUAUAGCUUAAAACUC
>170082.1 Mus setulosus
GAGAACUACUAGCUACAGCUUAAAACUC
>170139.1 Muscardinus avellanarius
GAGAACUACAAGCCAAAGCUAAAAACUC
>170227.1 Mustela nivalis
GAGAACUACUAGCAAUAGCUUAAAACUC
>170230.1 Mustela putorius
GAGAACUACUAGCAAUAGCUUAAAACUC
>170235.1 Neovison vison
GAGAACUACUAGCAACAGCCUAAAACUC
>172837.1 Myoictis melas
GAGAACUACUAGCCACUGCUUAAAACUC
>172841.1 Myoictis wallacei
GAGAACUACUAGCCACUGCUUAAAACUC
>172986.1 Glis glis
GAGUACUACAAGCCACAGCUCAAAACUC
>172987.1 Glis glis
GAGUACUACAAGCCACAGCUCAAAACUC
>176797.1 Neophascogale lorentzii
GAGAACUACAAGCCACUGCUUAAAACUC
>177465.1 Nesomys rufus
GAGAACUACUAGCUACAGCUUAAAACUC
>178128.1 Ningaui ridei
GAGAACUACGAGCCACUGCUUAAAACUC
>178129.1 Ningaui timealeyi
GAGAACUACGAGCCACUGCUUAAAACUC
>178130.1 Ningaui yvonnae
GAGAACUACGAGCCACUGCUUAAAACUC
>178650.1 Niviventer cremoriventer
GAGAACUACUAGCUACAGCUUAAAACUC
>180152.1 Notopteris macdonaldi
GAGUACUACUAGCAACAGCUUAAAACUC
>180157.1 Notoryctes typhlops
GAGAACUACAAGCGAGCGCUUAAAACUC
>180468.1 Nyctimene albiventer
GAGUACUACUAGCAACAGCUUAAAACUC
>180478.1 Nyctimene robinsoni
GAGUACUACUAGCAACAGCUUAAAACUC
>181753.1 Odocoileus virginianus
GAGUACUACCGGCAAUAGCUUAAAACUC
>182935.1 Onychogalea unguifera
GAGAACUACUAGCUAACGCUUAAAACUC
>184928.1 Ornithorhynchus anatinus
GAGAACUACUAGCAACAGCUUAAAACUC
>185359.1 Orycteropus afer
GAGAACUACAAGCAACAGCUUAAAACUC
>185361.1 Orycteropus afer
GAGAACUACAAGCAACAGCUUAAAACUC
>185377.1 Oryctolagus cuniculus
GAGAACUACAAGCCAAAGCUUAAAACUC
>185385.1 Oryx gazella
GAGUACUACCGGCAACGGCCCAAAACUC
>187081.1 Ovis aries
GAGUACUACC-GCAACAGCCCGAAACUC
>188919.1 Pan paniscus
GAACACUACGAGCCACAGCUUAAAACUC
>188990.1 Pan troglodytes
GAACACUACGAGCCACAGCUUAAAACUC
>188991.1 Pan troglodytes
GAACACUACGAGCCACAGCUUAAAACUC
>188992.1 Pan troglodytes
GAACACUACGAGCCACAGCUUAAAACUC
>188993.1 Pan troglodytes
GAACACUACGAGCCACAGCUUAAAACUC
>189529.1 Panthera leo
GAGAACUACUAGCAACAGCUUAAAACUC
>189562.1 Panthera tigris
GAGAACUACUAGCAACAGCUUAAAACUC
>189995.1 Papio hamadryas
GAAUACUACAAGCAACAGCUUAAAACUC
>191257.1 Pseudantechinus bilarni
GAGAACUACUAGCCACUGCUUAAAACUC
>193129.1 Pecari tajacu
GAGUACUACUAGCAACAGCCUAAAACUC
>195844.1 Petrogale concinna
GAGAACUACUAGCCAACGCUUAAAACUC
>195856.1 Perameles nasuta
GAGAACUACUAGCAAAUGCUUAAAACUC
>195857.1 Perameles nasuta
GAGAACUACUAGCAAAUGCUUAAAACUC
>196521.1 Peromyscus leucopus
GAGAACUACUGGCUACCGCUUAAAACUC
>196794.1 Peroryctes raffrayana
GAGAACUACUAGCAAAUGCUUAAAACUC
>198672.1 Phalanger gymnotis
GAGAACUACUAGCCAACGCUUAAAACUC
>198673.1 Phalanger lullulae
GAGAACUACUAGCCAACGCUUAAAACUC
>198674.1 Phalanger maculatus
GAGAACUACUAGCCAACGCUUAAAACUC
>198675.1 Phalanger maculatus
GAGAACUACUAGCCAACGCUUAAAACUC
>198677.1 Phalanger orientalis
GAGAACUACUAGCCAACGCUUAAAACUC
>198840.1 Phascogale calura
GAGAACUACAAGCUACCGCUUAAAACUC
>198843.1 Phascogale tapoatafa
GAGAACUACAAGCCACUGCUUAAAACUC
>198852.1 Phascolarctos cinereus
GAGAACUACUAGCCAGCGCUUAAAACUC
>198885.1 Phascolosorex dorsalis
GAGAACUACAAGCUACUGCUUAAAACUC
>200193.1 Phoca vitulina
GAGAACUACUAGCAACAGCUUAAAACUC
>206791.1 Pithecia pithecia
GAACACUACAAGCAACAGCUUGAAAUUC
>207550.1 Planigale gilesi
GAGAACUACGAGCCACCGCUUAAAACUC
>207551.1 Planigale ingrami
GAGAACUACGAGCCACCGCUUAAAACUC
>207572.1 Planigale tenuirostris
GAGAACUACGAGCCAUCGCUUAAAACUC
>213526.1 Pongo pygmaeus
GAACACUACGAGCCACAGCUUAAAACUC
>213537.1 Pongo abelii
GAACACUACGAGCCACAGCUUAAAACUC
>215104.1 Potorous longipes
GAGAACUACUAGCUAUUGCUUAAAACUC
>215836.1 Procavia capensis
GAGAACUACCAGCAAAAGCUAAAAACUC
>215991.1 Procyon lotor
GAGAACUACUAGCAACAGCUUAAAACUC
>217623.1 Pseudantechinus macdonnellensis
GAGAACUACUAGCCACUGCUUAAAACUC
>217627.1 Pseudantechinus woolleyae
GAGAACUACUAGCCACUGCUUAAAACUC
>218657.1 Pseudochirops cupreus
GGGAACUACUAGCUACUGCUUAAAACCC
>223316.1 Pteralopex atrata
GAGUACUACUAGCAACAGCUUAAAACUC
>223499.1 Pteropus admiralitatum
GAGUACUACUAGCAACAGCUUAAAACUC
>223514.1 Pteropus hypomelanus
GAGUACUACUAGCAACAGCUUAAAACUC
>224174.1 Puma concolor
GAGAACUACUAGCAACAGCUUAAAACUC
>227483.1 Rattus norvegicus
GAGAACUACUAGCUACAGCUUAAAACAC
>227485.1 Rattus norvegicus
GAGAACUACUAGCUACAGCUUAAAACUC
>227494.1 Rattus rattus
GAGAACUACUAGCUACAGCUUAAAACUC
>228641.1 Rhinoceros unicornis
GAGUACUACUAGCAACAGCCUAAAACUC
>232891.1 Rhyncholestes raphanurus
GAGAACUACUAGCAACUGCUUAAAACUC
>234326.1 Rousettus amplexicaudatus
GAGCACUACUAGCAACAGCUUAAAACUC
>234328.1 Myonycteris angolensis
GAGCACUACUAGCAACAGCUUGAAACUC
>235903.1 Saguinus geoffroyi
GAAUAUUACAAGCAACAGCUUGAAACUC
>235904.1 Saguinus oedipus
GAAUAUUACAAGCAACAGCUUGAAACUC
>235913.1 Saimiri sciureus
GAACACUACAAGCAACAGCUUGAAACUC
>237800.1 Scalopus aquaticus
GAGAACUACUAGCAAUAGCUUAAAACUC
>239876.1 Sciurus aestuans
GAGAACUACUAGCCACUGCUUAAAACUC
>243407.1 Setonix brachyurus
GAGAACUACUAGCCAACGCUUAAAACUC
>245942.1 Sminthopsis aitkeni
AAGAACUACGAGUCACUGCUUAAAACUC
>245943.1 Sminthopsis archeri
GAGAACUACGAGCUCCUGCUUAAAACUC
>245944.1 Sminthopsis bindi
GAGAACUACGAGCAACUGCUUAAAACUC
>245948.1 Sminthopsis crassicaudata
GAGAACUACGAGUCACUGCUUAAAACUC
>245951.1 Sminthopsis dolichura
GAGAACUACGAGUCACUGCUUAAAACUC
>245953.1 Sminthopsis douglasi
GAGAACUACGAGCAACUGCUUAAAACUC
>245958.1 Sminthopsis gilberti
GAGAACUACGAGUCACUGCUUAAAACUC
>245959.1 Sminthopsis granulipes
GAGAACUACGAGUCACUGCUUAAAACUC
>245960.1 Sminthopsis griseoventer
GAGAACUACGAGCUACUGCUUAAAACUC
>245961.1 Sminthopsis hirtipes
GAGAACUACGAGCCACUGCUUAAAACUC
>245963.1 Sminthopsis leucopus ferruginifrons
GAGAACUAUGAGUCACUGCUUAAAACUC
>245964.1 Sminthopsis leucopus leucopus
GAGAACUAUGAGUCACUGCUUAAAACUC
>245965.1 Sminthopsis longicaudata
GAGAACUAUGAGUCACUGCUUAAAACUC
>245966.1 Sminthopsis macroura
GAGAACUACGAGCCACCGCUUAAAACUC
>245984.1 Sminthopsis murina
GAGAACUACGAGCCACUGCUUAAAACUC
>245988.1 Sminthopsis ooldea
GAGAACUACGAGUCACUGCUUAAAACUC
>245989.1 Sminthopsis psammophila
GAGAACUACGAGUCACUGCUUAAAACUC
>245993.1 Sminthopsis virginiae nitela
GAGAACUACGAGUCACUGCUUAAAACUC
>245996.1 Sminthopsis virginiae rufigenis
GAGAACUACGAGUCACUGCUUAAAACUC
>245997.1 Sminthopsis virginiae virginiae
GAGAACUACGAGUCACUGCUUAAAACUC
>245998.1 Sminthopsis youngsoni
GAGAACUACGAGUCACUGCUUAAAACUC
>246799.1 Solenodon paradoxus
GAGUACUACUAGCCACAGCUUAAAACUC
>247114.1 Sorex palustris
GAGAACUACUAGCAAUAGCUUAAAACUC
>249912.1 Spilocuscus rufoniger
GAGGACUACUAGCCAACGCUUGAAACUC
>249915.1 Spilogale putorius
GAGAACUACUAGCAACAGCCUGAAACUC
>253361.1 Stenella coeruleoalba
GAAUACUAUCGGCAACAGCCA-AAACUC
>260454.1 Syconycteris australis
GAGUACUACUAGCAACAGCUUAAAACUC
>260528.1 Sylvilagus audubonii
GAGAACUACAAGCCAGAGCUUAAAACUC
>264547.1 Tapirus pinchaque
GAGUACUACUAGCAACAGCUUAAAACUC
>264849.1 Tarsius syrichta
GAGUACUACAAGCAACAGCUUAAAACUC
>264895.1 Gerbilliscus kempi gambiana
GAGAACUACGAGCCACCGCUUAAAACUC
>269256.1 Thoopterus nigrescens
GAGAACUACUAGCAACAGCUUAAAACUC
>269596.1 Thylogale stigmatica
GAGAACUACUAGCCAACGCUUAAAACUC
>272893.1 Tragelaphus imberbis
GAGUACUACUAGCAACAGCUUAAAACUC
>272948.1 Tragulus napu
GAGAACUACUAGCCACAGCUUAAAACUC
>274024.1 Trichechus manatus
GAGAACUACUAGCAAAGGCUUAAAACUC
>276078.1 Trichosurus vulpecula
GAGAACUACUAGCCAGCGCUUAAAACUC
>280018.1 Tupaia tana
GAGUACUACGUGCCACAGCCCAAAACUC
>281605.1 Uranomys ruddi
GAGAACUACGAGCCACAGCUUAAAACUC
>282267.1 Ursus americanus
GAGAACUACUAGCAACAGCUUAAAACUC
>282273.1 Ursus arctos
GAGAACUACUAGCAACAGCUUAAAACUC
>286427.1 Vombatus ursinus
GAGAACUACUAGCCACCGCUUAAAACUC
>286490.1 Vulpes vulpes
GAGAACUACUAGCAACAGCUUAAAACUC
>286635.1 Wallabia bicolor
GAGAACUACUAGCCAACGCUUAAAACUC
>289276.1 Zalophus californianus
GAGAACUACUAGCAAUAGCUUAAAACUC
>472474.1 Dendrohyrax dorsalis
GAGAACUACUAGCAAUAGCUAAAAACUC
Loop 1 Internal loop
Column numbers: 3-10, 21-26
GUACUACU*UAAAAC
ACACUGCA*UAAAAC
GAACUACU*UAAAAC
GUACUAUC*A-AAAC
ACACUACG*UAAAAC
GUACUACU*UAAAAC
GUACUACU*UAAAAC
GAACUACU*UAAAAC
GUACUACU*UAAAAC
GAACUACU*UAAAAC
GUACUACC*CAAAAC
GAACUACA*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
ACACUACA*UGAAAC
ACACUACA*UGAAAU
GAACUACU*UAAAAC
GAACUACG*UAAAAC
GAACUACA*UAAAAC
GAACUACA*UAAAAC
GAACUACA*UAAAAC
GAACUACA*UAAAAC
GAACUACA*UAAAAC
GAACUACA*UAAAAC
GAACUACA*UAAAAC
GAACUACA*UAAAAC
GAACUACA*UAAAAC
GAACUACA*UAAAAC
GAACUACA*UAAAAC
GAACUACG*UAAAAC
GAACUACG*UAAAAC
GUACUACU*UAAAAC
AUACUACA*UGAAAC
GUACUACG*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GUACUACU*UAAAAC
GAACUACG*UAAAAC
ACACUACA*UGAAAC
GUACUACU*UAAAAC
GUACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GGACUACU*UAAAAC
GUACUACU*UAAAAC
GUACUACC*UAAAAC
ACACUACA*UGAAAC
GUACUACU*UGAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
ACACUACA*UGAAAC
ACACUACA*UGAAAU
ACACUACA*UGAAAU
ACACUACA*UGAAAC
GAACUACU*UAAAAC
GUACUACU*UAAAAC
GGACUACU*UAAAAC
GGACUACU*UAAAAC
GUACUACC*CGAAAC
ACACUACA*UAAAAC
GUACUACC*UAAAAC
GUACUACU*UAAAAC
GUACUACU*UAAAAC
GUACUACC*UAAAAC
GAACUACU*UGAAAC
GUACUACU*UAAAAC
ACACUACA*UGAAAU
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GUACUACC*CAAAAC
GAACUACU*UAAAAC
GUACUACA*CAAAAC
GAACUACU*UAAAAC
GUACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GCACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GUACUACA*CAAAAC
GAACUACU*UAAAAC
GAACUACU*AAAAAC
GUACUACU*UAAAAC
GUACUACG*UAAAAC
GAACUACA*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACG*CAAAAC
GAACUACU*UAAAAC
GCACUACU*UAAAAC
GCACUACU*UAAAAC
GUACUACC*AGAAAC
GUACUACU*UAAAAC
GUACUACU*UAAAAC
GUACUACU*UAAAAC
GAACUACG*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GUACUACC*UAAAAC
GAACUACU*UAAAAC
GUACUACU*UAAAAC
ACACUACG*UAAAAC
ACACUACG*UAAAAC
GAACUAUA*AAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GUACUACU*UAAAAC
GUACUACU*UAAAAC
GUACUACU*UAAAAC
GUACUACC*UAAAAC
ACACUACG*UAAAAC
ACACUACG*UAAAAC
ACACUACG*UAAAAC
ACACUACG*UAAAAC
ACACUACG*UAAAAC
GAACUACU*UAAAAC
GUACUACC*UAAAAC
ACACUACG*UAAAAA
ACACUACG*UAAAAA
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GUACUACC*UAAAAC
GAACUACU*UAAAAC
ACACUACA*UGAAAC
GUACUACU*UAAAAC
GCACUACA*UAAAAC
ACACUACA*UGAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GGACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UGAAAC
GUACUACC*UAAAAC
GAACUACU*UAAAAC
GUACUACU*UGAAAC
GAACUACU*UAAAAC
GUACUACG*UAAAAC
GUACUACU*UAAAAC
GAACUACU*UAAAAC
GUACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GUACUACU*UAAAAC
GAACUACU*UAAAAC
AAACUACU*UAAAAC
GUACUACC*UAAAAC
GAACUACA*UAAAAC
GAACUACA*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACA*AAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GUACUACA*CAAAAC
GUACUACA*CAAAAC
GAACUACA*UAAAAC
GAACUACU*UAAAAC
GAACUACG*UAAAAC
GAACUACG*UAAAAC
GAACUACG*UAAAAC
GAACUACU*UAAAAC
GUACUACU*UAAAAC
GAACUACA*UAAAAC
GUACUACU*UAAAAC
GUACUACU*UAAAAC
GUACUACC*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACA*UAAAAC
GAACUACA*UAAAAC
GAACUACA*UAAAAC
GUACUACC*CAAAAC
GUACUACC*CGAAAC
ACACUACG*UAAAAC
ACACUACG*UAAAAC
ACACUACG*UAAAAC
ACACUACG*UAAAAC
ACACUACG*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
AUACUACA*UAAAAC
GAACUACU*UAAAAC
GUACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACA*UAAAAC
GAACUACA*UAAAAC
GAACUACU*UAAAAC
GAACUACA*UAAAAC
GAACUACU*UAAAAC
ACACUACA*UGAAAU
GAACUACG*UAAAAC
GAACUACG*UAAAAC
GAACUACG*UAAAAC
ACACUACG*UAAAAC
ACACUACG*UAAAAC
GAACUACU*UAAAAC
GAACUACC*AAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GUACUACU*UAAAAC
GUACUACU*UAAAAC
GUACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GUACUACU*UAAAAC
GAACUACU*UAAAAC
GCACUACU*UAAAAC
GCACUACU*UGAAAC
AUAUUACA*UGAAAC
AUAUUACA*UGAAAC
ACACUACA*UGAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACG*UAAAAC
GAACUACG*UAAAAC
GAACUACG*UAAAAC
GAACUACG*UAAAAC
GAACUACG*UAAAAC
GAACUACG*UAAAAC
GAACUACG*UAAAAC
GAACUACG*UAAAAC
GAACUACG*UAAAAC
GAACUACG*UAAAAC
GAACUAUG*UAAAAC
GAACUAUG*UAAAAC
GAACUAUG*UAAAAC
GAACUACG*UAAAAC
GAACUACG*UAAAAC
GAACUACG*UAAAAC
GAACUACG*UAAAAC
GAACUACG*UAAAAC
GAACUACG*UAAAAC
GAACUACG*UAAAAC
GAACUACG*UAAAAC
GUACUACU*UAAAAC
GAACUACU*UAAAAC
GGACUACU*UGAAAC
GAACUACU*UGAAAC
AUACUAUC*A-AAAC
GUACUACU*UAAAAC
GAACUACA*UAAAAC
GUACUACU*UAAAAC
GUACUACA*UAAAAC
GAACUACG*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GUACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GUACUACG*CAAAAC
GAACUACG*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*UAAAAC
GAACUACU*AAAAAC
Click on headings to reorder table
- View All Results for this Loop
| # | Matching motif group | Acceptance Rate | Mean Cutoff Score | Full Edit Distance | Interior Edit Distance | Basepair Diagram and most common annotation |
|---|---|---|---|---|---|---|
| 1 |
Motif group IL_95652.6
Basepair signature: |
99.02 | 35.28 | 3 | 2 |
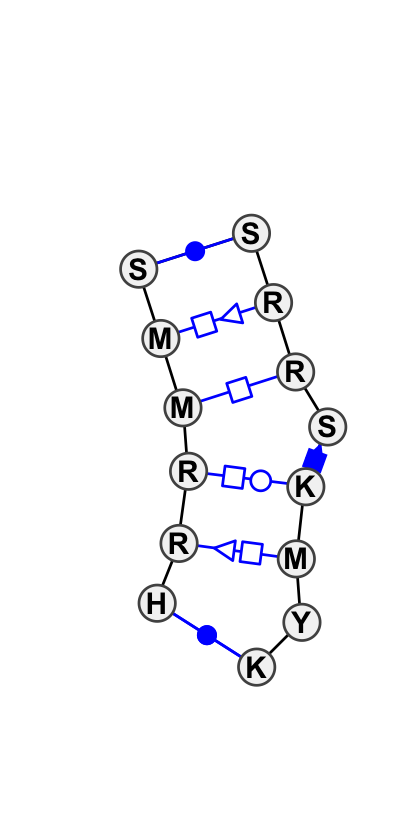
|
| 2 |
Motif group IL_73656.1
Basepair signature: |
22.15 | -11.84 | 6 | 4 |
|
| 3 |
Motif group IL_06584.1
Basepair signature: |
19.87 | -10.09 | 8 | 4 |

|
| 4 |
Motif group IL_48045.1
Basepair signature: |
6.51 | -17.39 | 7 | 5 |
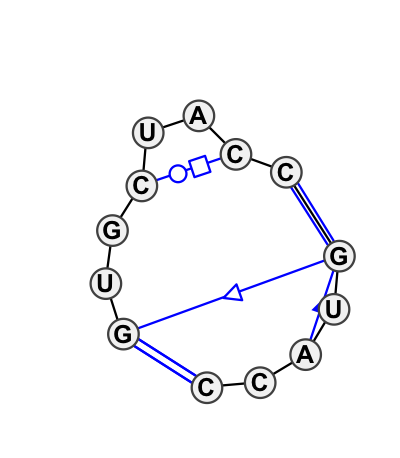
|
| 5 |
Motif group IL_62296.1
Basepair signature: |
4.89 | -27.09 | 7 | 5 |

|
| 6 |
Motif group IL_91857.1
Basepair signature: |
2.93 | -24.43 | 8 | 5 |

|
| 7 |
Motif group IL_29058.1
Basepair signature: |
0.98 | -29.20 | 7 | 4 |

|
| 8 |
Motif group IL_15702.2
Basepair signature: |
0.65 | -30.39 | 7 | 4 |

|
| 9 |
Motif group IL_53635.6
Basepair signature: |
0.65 | -43.07 | 6 | 4 |
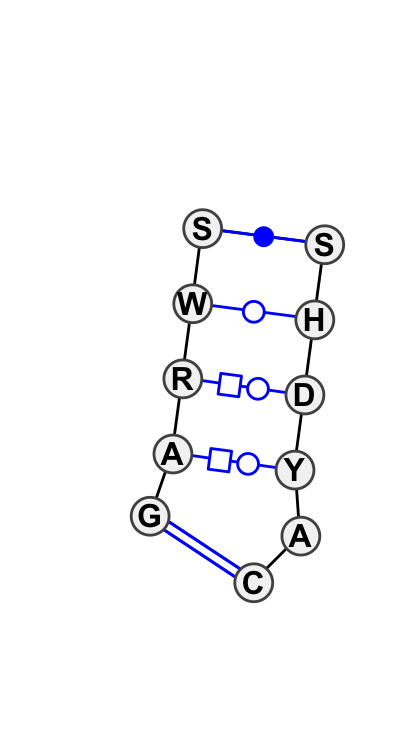
|
| 10 |
Motif group IL_63952.1
Basepair signature: |
0.33 | -22.59 | 5 | 4 |
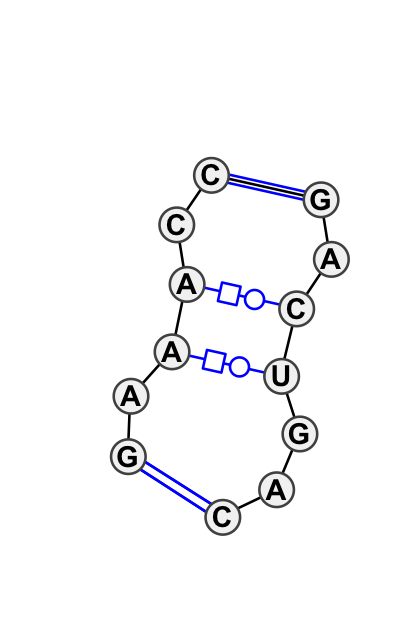
|
Loop 2 Hairpin loop
Column numbers: 13-18
CAACUG
CAACAG
CAAUAG
CAACAG
CCACAG
CAACUG
CAACUG
CCACAG
CAACAG
CCAUAG
CAACGG
CCAAUG
CAACAG
CAAUAG
CAACAG
CAACAG
CAAUAG
CCACCG
CCACUG
CCACUG
CCACUG
CCACUG
CCACUG
CCACUG
CCACUG
CUACUG
CCACUG
CUACUG
CCACUG
CCACUG
CCACUG
CAACUG
CCACAG
CAACAG
CAACAG
CAACAG
CCAAAG
CCACAG
CAACAG
CAACAG
CAACAG
CAACAG
CCAAUG
CAACAG
CAACAG
CAACAG
CAACAG
CAAUAG
CCAGCG
CAAUCG
CCACAG
CAACAG
CAACAG
CAACAG
CAACUG
CAACAG
CAAUAG
CAAUAG
CAACAG
CAAUAG
CAACAG
CAACAG
CAACAG
CAAUAG
CAAAUG
CAAUAG
CAACAG
CCACAG
CCACAG
CCACCG
CAACGG
CAACAG
CCACAG
CCACUG
CAACAG
CCACUG
CCACUG
CCACUG
CCACUG
CCACUG
CCAAUG
CCAACG
CAAUUG
CAAUAG
CCAGCG
CCAGCG
CUAAUG
CCAUAG
CAAAAG
CAAA-G
CAAAUG
CAACAG
CCACUG
CCAGAG
CCAGAG
CCAGAG
CCACAG
CAAUAG
CAACAG
CAACAG
CAAGGG
CAACAG
CAACAG
CAACAG
CUACAG
CAAUAG
CAAUAG
CAACAG
CCAACG
CAAUUG
CCACAG
CCACAG
CAACAG
CAACAG
CAACAG
CAAUAG
CAACAG
CAACAG
CAACAG
CCACAG
CCACAG
CCACAG
CCACAG
CCACAG
CAACAG
CAAUAG
CAACAG
CAACAG
CCAUAG
CAAUCG
CAAAUG
CAAUAG
CCAACG
CAAUAG
CAACAG
CAACAG
CAACAG
CCACAG
CAACAG
CCAGAG
CCAGAG
CCAGAG
CCAUUG
CUACAG
CCAAUG
CCAAUG
CCAAUG
CCAGUG
CCACAG
CAAAUG
CAACAG
CCAGAG
CAAUAG
CCACAG
CAACAG
CAACAG
CAACAG
CAACAG
CAACAG
CCACAG
CAAAUG
CCACAG
CAACAG
CAAAAG
CAAUAG
CUACUG
CAAUAG
CUACUG
CUACUG
CCAUAG
CCACAG
CUACAG
CCAUAG
CCAUAG
CCAUAG
CCAUAG
CCAUAG
CUACAG
CUAUAG
CUAUAG
CUACAG
CCAAAG
CAAUAG
CAAUAG
CAACAG
CCACUG
CCACUG
CCACAG
CCACAG
CCACUG
CUACAG
CCACUG
CCACUG
CCACUG
CUACAG
CAACAG
CGAGCG
CAACAG
CAACAG
CAAUAG
CUAACG
CAACAG
CAACAG
CAACAG
CCAAAG
CAACGG
CAACAG
CCACAG
CCACAG
CCACAG
CCACAG
CCACAG
CAACAG
CAACAG
CAACAG
CCACUG
CAACAG
CCAACG
CAAAUG
CAAAUG
CUACCG
CAAAUG
CCAACG
CCAACG
CCAACG
CCAACG
CCAACG
CUACCG
CCACUG
CCAGCG
CUACUG
CAACAG
CAACAG
CCACCG
CCACCG
CCAUCG
CCACAG
CCACAG
CUAUUG
CAAAAG
CAACAG
CCACUG
CCACUG
CUACUG
CAACAG
CAACAG
CAACAG
CAACAG
CUACAG
CUACAG
CUACAG
CAACAG
CAACUG
CAACAG
CAACAG
CAACAG
CAACAG
CAACAG
CAAUAG
CCACUG
CCAACG
UCACUG
CUCCUG
CAACUG
UCACUG
UCACUG
CAACUG
UCACUG
UCACUG
CUACUG
CCACUG
UCACUG
UCACUG
UCACUG
CCACCG
CCACUG
UCACUG
UCACUG
UCACUG
UCACUG
UCACUG
UCACUG
CCACAG
CAAUAG
CCAACG
CAACAG
CAACAG
CAACAG
CCAGAG
CAACAG
CAACAG
CCACCG
CAACAG
CCAACG
CAACAG
CCACAG
CAAAGG
CCAGCG
CCACAG
CCACAG
CAACAG
CAACAG
CCACCG
CAACAG
CCAACG
CAAUAG
CAAUAG
Click on headings to reorder table
- View All Results for this Loop
| # | Matching motif group | Acceptance Rate | Mean Cutoff Score | Full Edit Distance | Interior Edit Distance | Basepair Diagram and most common annotation |
|---|---|---|---|---|---|---|
| 1 |
Motif group HL_48116.3
Basepair signature: |
94.14 | 55.67 | 1 | 1 |
|
| 2 |
Motif group HL_19626.4
Basepair signature: |
93.81 | 30.74 | 2 | 1 |
|
| 3 |
Motif group HL_35865.1
Basepair signature: |
90.23 | 31.37 | 3 | 1 |
|
| 4 |
Motif group HL_11376.1
Basepair signature: |
86.97 | 22.56 | 3 | 2 |
|
| 5 |
Motif group HL_79956.2
Basepair signature: |
79.80 | 34.19 | 2 | 2 |
|
| 6 |
Motif group HL_27271.4
Basepair signature: |
75.24 | 27.76 | 2 | 1 |
|
| 7 |
Motif group HL_66677.2
Basepair signature: |
73.94 | 28.17 | 2 | 2 |
|
| 8 |
Motif group HL_79038.1
Basepair signature: |
71.34 | -0.40 | 4 | 2 |

No annotation |
| 9 |
Motif group HL_14536.1
Basepair signature: |
71.01 | -0.32 | 4 | 2 |
|
| 10 |
Motif group HL_74411.3
Basepair signature: |
70.03 | 24.74 | 2 | 1 |
|