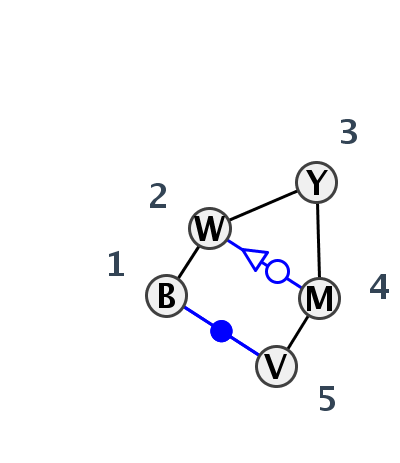
Motif HL_22584.1 Version HL_22584.1 of this group appears in releases 3.87 to 3.87
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 1-5 | 2-3 | 2-4 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HL_6OL3_001 | 6OL3 | 0.4783 | 2 | Other HL | C | VA RNA | G | 64 | A | 66 | C | 68 | C | 69 | C | 70 | cWW | ntSW | |
| 2 | HL_8DK7_001 | 8DK7 | 0.1308 | 2 | Fab BL3-6 binding hairpin with tSW | C | RNA (34-MER) | G | 13 | A | 15 | C | 17 | A | 18 | C | 19 | cWW | tSW | |
| 3 | HL_3IVK_003 | 3IVK | 0.1583 | 2 | Fab BL3-6 binding hairpin with tSW | M | class I ligase product | G | 59 | A | 61 | C | 63 | A | 64 | C | 65 | cWW | tSW | |
| 4 | HL_8VM8_003 | 8VM8 | 0.1514 | 2 | Fab BL3-6 binding hairpin with tSW | R | Enterovirus 5' cloverleaf cis-acting replication element | G | 63 | A | 65 | C | 67 | A | 68 | C | 69 | cWW | tSW | |
| 5 | HL_4KZD_001 | 4KZD | 0.1507 | 2 | Fab BL3-6 binding hairpin with tSW | R | RNA (84-MER) | G | 37 | A | 39 | C | 41 | A | 42 | C | 43 | cWW | tSW | |
| 6 | HL_6DB9_001 | 6DB9 | 0.1466 | 2 | Fab BL3-6 binding hairpin with tSW | R | RNA (60-MER) | G | 23 | A | 25 | C | 27 | A | 28 | C | 29 | cWW | tSW | |
| 7 | HL_8D29_002 | 8D29 | 0.1171 | 2 | Fab BL3-6 binding hairpin with tSW | F | RNA (34-MER) | G | 13 | A | 15 | C | 17 | A | 18 | C | 19 | cWW | tSW | |
| 8 | HL_8D29_003 | 8D29 | 0.1186 | 2 | Fab BL3-6 binding hairpin with tSW | J | RNA (34-MER) | G | 13 | A | 15 | C | 17 | A | 18 | C | 19 | cWW | tSW | |
| 9 | HL_8VM9_006 | 8VM9 | 0.1200 | 2 | Fab BL3-6 binding hairpin with tSW | R | Enterovirus 5' cloverleaf cis-acting replication element | G | 57 | A | 59 | C | 61 | A | 62 | C | 63 | cWW | tSW | |
| 10 | HL_8T29_001 | 8T29 | 0.1053 | 2 | Fab BL3-6 binding hairpin with tSW | R | RNA (90-MER) | G | 35 | A | 37 | C | 39 | A | 40 | C | 41 | cWW | tSW | |
| 11 | HL_8VMB_001 | 8VMB | 0.1018 | 2 | Fab BL3-6 binding hairpin with tSW | R | Enterovirus 5' cloverleaf cis-acting replication element | G | 18 | A | 20 | C | 22 | A | 23 | C | 24 | cWW | ntSW | |
| 12 | HL_8DP3_001 | 8DP3 | 0.0000 | 2 | Fab BL3-6 binding hairpin with tSW | R | Enterovirus 5' cloverleaf cis-acting replication element | G | 20 | A | 22 | C | 24 | A | 25 | C | 26 | cWW | tSW | |
| 13 | HL_8VMA_003 | 8VMA | 0.0916 | 2 | Fab BL3-6 binding hairpin with tSW | C | Enterovirus 5' cloverleaf cis-acting replication element | G | 60 | A | 62 | C | 64 | A | 65 | C | 66 | cWW | tSW | |
| 14 | HL_7MLX_002 | 7MLX | 0.0772 | 2 | Fab BL3-6 binding hairpin with tSW | R | RNA (65-MER) | G | 40 | A | 42 | C | 44 | A | 45 | C | 46 | cWW | tSW | |
| 15 | HL_8D29_001 | 8D29 | 0.0819 | 2 | Fab BL3-6 binding hairpin with tSW | C | RNA (34-MER) | G | 13 | A | 15 | C | 17 | A | 18 | C | 19 | cWW | tSW | |
| 16 | HL_6X5M_002 | 6X5M | 0.1063 | 2 | Fab BL3-6 binding hairpin with tSW | r | ggPAN RNA (39-MER) | G | 14 | A | 16 | C | 18 | A | 19 | C | 20 | cWW | tSW | |
| 17 | HL_7SZU_001 | 7SZU | 0.1133 | 2 | Fab BL3-6 binding hairpin with tSW | R | RNA aptamer | G | 35 | A | 37 | C | 39 | A | 40 | C | 41 | cWW | tSW | |
| 18 | HL_8UIW_001 | 8UIW | 0.0896 | 2 | Fab BL3-6 binding hairpin with tSW | N | azaaromatic riboswitch aptamer (yjdF RNA) | G | 29 | A | 31 | C | 33 | A | 34 | C | 35 | cWW | tSW | |
| 19 | HL_7A0S_061 | 7A0S | 0.4909 | 2 | LSU A loop | X | LSU rRNA | C | 2530 | U | 2531 | U | 2534 | C | 2535 | G | 2536 | cWW | ||
| 20 | HL_4WF9_058 | 4WF9 | 0.4802 | 2 | LSU A loop | X | LSU rRNA | C | 2578 | U | 2579 | U | 2582 | C | 2583 | G | 2584 | cWW | ||
| 21 | HL_5TBW_063 | 5TBW | 0.5458 | 2 | 1 | LSU rRNA | U | 2920 | U | 2921 | U | 2924 | C | 2925 | A | 2926 | cWW | |||
| 22 | HL_8C3A_064 | 8C3A | 0.5641 | 2 | 1 | LSU rRNA | U | 2892 | U | 2893 | U | 2896 | C | 2897 | A | 2898 | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GAAACAC | 17 |
| CUGUUCG | 2 |
| UUGUUCA | 2 |
| GAACCCC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| AAACA | 17 |
| UGUUC | 4 |
| AACCC | 1 |
Release history
| Release | 3.87 |
|---|---|
| Date | 2024-08-14 |
| Status | New id, 2 parents |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- Fab BL3-6 binding hairpin with tSW (17)
- LSU A loop (2)
- (2)
- Other HL (1)
- Basepair signature
- cWW-tSW-F
- Heat map statistics
- Min 0.03 | Avg 0.27 | Max 0.95
Coloring options: