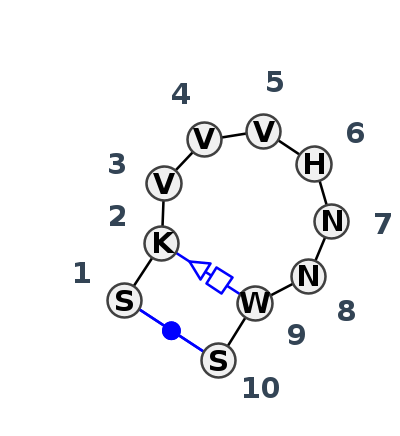
Motif HL_38649.1 Version HL_38649.1 of this group appears in releases 3.89 to 3.99
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 1-10 | 2-8 | 2-9 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HL_4RMO_001 | 4RMO | 0.7641 | 2 | B | RNA (45-MER) | C | 11 | U | 12 | C | 15 | C | 16 | G | 17 | A | 18 | U | 19 | A | 20 | U | 21 | G | 22 | cWW | tWH | |
| 2 | HL_2A64_006 | 2A64 | 0.2593 | 2 | A | RNase P class B | C | 313 | G | 314 | G | 317 | C | 318 | A | 319 | U | 320 | G | 321 | C | 322 | A | 323 | G | 324 | cWW | ntSH | |
| 3 | HL_3HHN_006 | 3HHN | 0.0000 | 2 | E | Class I ligase ribozyme, self-ligation product | G | 1 | G | 2 | C | 5 | A | 6 | C | 7 | U | 8 | A | 9 | U | 10 | A | 11 | C | 12 | cWW | tSH | |
| 4 | HL_3IVK_001 | 3IVK | 0.1330 | 2 | M | class I ligase product | G | 1 | G | 2 | C | 5||A | A | 6 | C | 7 | U | 8 | A | 9 | U | 10 | A | 11 | C | 12 | cWW | tSH | |
| 5 | HL_7MLX_001 | 7MLX | 0.5768 | 2 | R | RNA (65-MER) | G | 11 | U | 12 | A | 15 | G | 16 | C | 17 | C | 18 | C | 19 | G | 20 | U | 21 | C | 22 | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GGAACACUAUAC | 2 |
| CUGACCGAUAUG | 1 |
| CGGCGCAUGCAG | 1 |
| GUGCAGCCCGUC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GAACACUAUA | 2 |
| UGACCGAUAU | 1 |
| GGCGCAUGCA | 1 |
| UGCAGCCCGU | 1 |
Release history
| Release | 3.89 | 3.90 | 3.91 | 3.92 | 3.93 | 3.94 | 3.95 | 3.96 | 3.97 | 3.98 | 3.99 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Date | 2024-10-09 | 2024-11-06 | 2024-12-04 | 2025-01-01 | 2025-01-29 | 2025-02-26 | 2025-03-26 | 2025-04-23 | 2025-05-21 | 2025-06-18 | 2025-07-16 |
| Status | New id, > 2 parents | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (5)
- Basepair signature
- cWW-tSH-F-F-F-F-F-F
- Heat map statistics
- Min 0.13 | Avg 0.46 | Max 0.92
Coloring options: