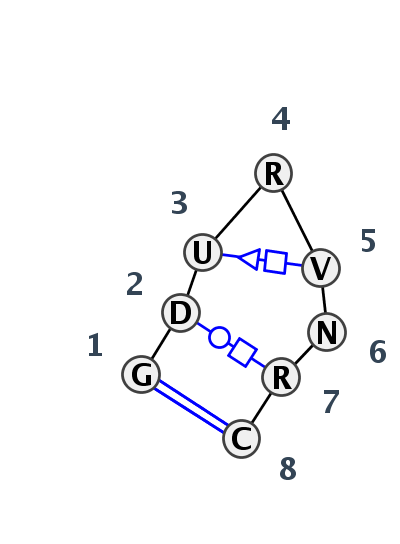
Motif HL_69023.1 Version HL_69023.1 of this group appears in releases 3.77 to 3.79
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 1-8 | 2-7 | 3-6 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HL_5X2G_003 | 5X2G | 0.5565 | 1 | T-loop related | B | sgRNA | G | 73 | U | 74 | U | 75 | A | 76 | C | 77 | A | 78 | A | 79 | C | 81 | cWW | tWH | |
| 2 | HL_7MLW_003 | 7MLW | 0.0000 | 1 | T-loop related | F | Guanidine-I riboswitch | G | 112 | A | 113 | U | 114 | A | 115 | A | 116 | A | 117 | A | 118 | C | 120 | cWW | tWH | tWH |
| 3 | HL_6DLR_003 | 6DLR | 0.0846 | 1 | T-loop related | A | Guanidine-I riboswitch | G | 82 | A | 83 | U | 84 | A | 85 | G | 86 | A | 87 | A | 88 | C | 90 | cWW | tWH | tWH |
| 4 | HL_5T83_003 | 5T83 | 0.0981 | 1 | T-loop related | A | Guanidine-I riboswitch | G | 77 | A | 78 | U | 79 | A | 80 | A | 81 | A | 82 | A | 83 | C | 85 | cWW | ntWH | tWH |
| 5 | HL_4V88_193 | 4V88 | 0.4656 | 1 | GNRA related | A6 | SSU rRNA | G | 330 | A | 331 | U | 332 | A | 333 | G | 334 | U | 335 | G | 336 | C | 338 | cWW | ||
| 6 | HL_6CZR_077 | 6CZR | 0.4743 | 1 | GNRA related | 1a | SSU rRNA | G | 255 | G | 256 | U | 257 | A | 258 | A | 259 | U | 260 | G | 261 | C | 263 | cWW | ||
| 7 | HL_4LFB_006 | 4LFB | 0.4780 | 1 | GNRA related | A | SSU rRNA | G | 259 | G | 260 | U | 261 | A | 262 | A | 263 | U | 264 | G | 265 | C | 267 | cWW | ||
| 8 | HL_5J7L_006 | 5J7L | 0.5508 | 1 | GNRA related | AA | SSU rRNA | G | 259 | G | 260 | U | 261 | A | 262 | A | 263 | C | 264 | G | 265 | C | 267 | cWW | ||
| 9 | HL_7RQB_036 | 7RQB | 0.7882 | 2 | 1A | LSU rRNA | G | 1389 | U | 1390 | U | 1391 | A | 1392 | A | 1393 | U | 1394 | A | 1395 | C | 1398 | cWW | tWH | ||
| 10 | HL_5J7L_168 | 5J7L | 0.7932 | 2 | DA | LSU rRNA | G | 1389 | U | 1390 | U | 1391 | A | 1392 | A | 1393 | U | 1394 | A | 1395 | C | 1398 | cWW | tWH | ||
| 11 | HL_4V9F_037 | 4V9F | 0.8039 | 2 | 0 | LSU rRNA | G | 1498 | U | 1499 | U | 1500 | A | 1501 | A | 1502 | U | 1503 | A | 1504 | C | 1507 | cWW | tWH | ||
| 12 | HL_4WF9_035 | 4WF9 | 0.8180 | 2 | X | LSU rRNA | G | 1426 | U | 1427 | U | 1428 | G | 1429 | A | 1430 | U | 1431 | A | 1432 | C | 1435 | cWW | tWH | ||
| 13 | HL_5TBW_035 | 5TBW | 0.8654 | 2 | 1 | LSU rRNA | G | 1599 | U | 1600 | U | 1601 | A | 1602 | A | 1603 | G | 1604 | A | 1605 | C | 1608 | cWW | tWH |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GUUAAUAUUC | 3 |
| GAUAAAAGC | 2 |
| GGUAAUGGC | 2 |
| GUUACAAUC | 1 |
| GAUAGAAGC | 1 |
| GAUAGUGGC | 1 |
| GGUAACGGC | 1 |
| GUUGAUAUUC | 1 |
| GUUAAGAUUC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| UUAAUAUU | 3 |
| AUAAAAG | 2 |
| GUAAUGG | 2 |
| UUACAAU | 1 |
| AUAGAAG | 1 |
| AUAGUGG | 1 |
| GUAACGG | 1 |
| UUGAUAUU | 1 |
| UUAAGAUU | 1 |
- Annotations
-
- (5)
- GNRA related (4)
- T-loop related (4)
- Basepair signature
- cWW-tWH-tSH-F-F
- Heat map statistics
- Min 0.08 | Avg 0.50 | Max 0.88
Coloring options: