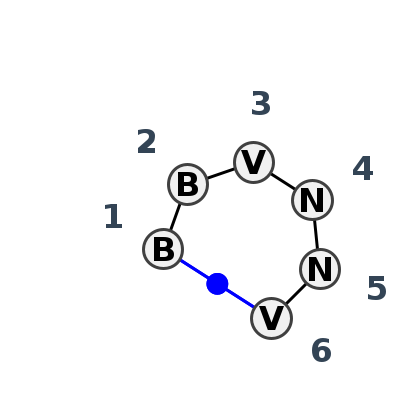
Motif HL_73355.1 Version HL_73355.1 of this group appears in releases 3.88 to 3.88
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 1-6 | 2-4 | 2-5 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HL_3OIN_001 | 3OIN | 0.6875 | 2 | C | RNA (14-mer) | C | 4 | U | 5 | A | 8 | A | 9 | C | 10 | G | 11 | cWW | |||
| 2 | HL_2A64_006 | 2A64 | 0.5714 | 6 | A | RNase P class B | C | 313 | G | 314 | G | 321 | C | 322 | A | 323 | G | 324 | cWW | ntSH | ||
| 3 | HL_4LX6_002 | 4LX6 | 0.1232 | 2 | Purine riboswitch | A | Purine riboswitch | G | 46 | C | 47 | C | 50 | C | 51 | A | 52 | C | 53 | cWW | ||
| 4 | HL_4XNR_002 | 4XNR | 0.0725 | 2 | Purine riboswitch | X | Purine riboswitch | G | 46 | U | 47 | C | 50 | U | 51 | A | 52 | C | 53 | cWW | ntWH | |
| 5 | HL_1Y26_002 | 1Y26 | 0.0595 | 2 | Purine riboswitch | X | Purine riboswitch | G | 46 | U | 47 | C | 50 | U | 51 | A | 52 | C | 53 | cWW | ntWH | |
| 6 | HL_1Y27_002 | 1Y27 | 0.0614 | 2 | Purine riboswitch | X | Purine riboswitch | G | 46 | U | 47 | C | 50 | U | 51 | A | 52 | C | 53 | cWW | ||
| 7 | HL_4FEN_002 | 4FEN | 0.0000 | 2 | Purine riboswitch | B | Purine riboswitch | G | 46 | U | 47 | C | 50 | U | 51 | A | 52 | C | 53 | cWW | ntWH | |
| 8 | HL_3RKF_008 | 3RKF | 0.1254 | 2 | Purine riboswitch | C | Purine riboswitch | G | 46 | U | 47 | C | 50 | U | 51 | A | 52 | C | 53 | cWW | ntWH | |
| 9 | HL_3LA5_002 | 3LA5 | 0.1578 | 2 | Purine riboswitch | A | Purine riboswitch | G | 46 | C | 47 | C | 50 | C | 51 | A | 52 | C | 53 | cWW | ||
| 10 | HL_7TZS_001 | 7TZS | 0.4934 | 1 | X | TPP riboswitch (THI element) | C | 27 | U | 28 | C | 30 | G | 31 | U | 32 | G | 33 | cWW | tWW | ||
| 11 | HL_5VPO_138 | 5VPO | 0.5401 | 2 | Pseudoknot geometry | XV | P-site ASL SufA6 | U | 32 | U | 33 | G | 35 | G | 36 | 1MG | 37 | A | 39 | ncWW | ||
| 12 | HL_6UFH_003 | 6UFH | 0.5918 | 2 | Pseudoknot geometry | A | RNA (167-MER) | G | 70 | U | 72 | G | 74 | G | 75 | C | 76 | C | 77 | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GUUUCUAC | 5 |
| GCUUCCAC | 2 |
| CUUCAACG | 1 |
| CGGCGCAUGCAG | 1 |
| CUGCGUG | 1 |
| UUCGGGA | 1 |
| GGUUGGCC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| UUUCUA | 5 |
| CUUCCA | 2 |
| UUCAAC | 1 |
| GGCGCAUGCA | 1 |
| UGCGU | 1 |
| UCGG(1MG)G | 1 |
| GUUGGC | 1 |
Release history
| Release | 3.88 |
|---|---|
| Date | 2024-09-11 |
| Status | New id, 2 parents |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- Purine riboswitch (7)
- (3)
- Pseudoknot geometry (2)
- Basepair signature
- cWW-F-F-F-F
- Heat map statistics
- Min 0.06 | Avg 0.41 | Max 0.87
Coloring options: