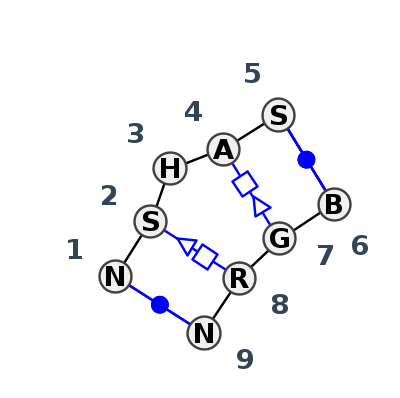
Motif IL_45790.1 Version IL_45790.1 of this group appears in releases 4.0 to 4.1
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | break | 6 | 7 | 8 | 9 | 1-9 | 2-8 | 4-7 | 5-6 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_2R8S_006 | 2R8S | 0.3247 | 1 | UAA/GAN variation | R | P4-P6 RNA RIBOZYME DOMAIN | C | 216 | C | 217 | A | 218 | A | 219 | G | 220 | * | U | 253 | G | 254 | A | 256 | G | 257 | cWW | tSH | tHS | cWW |
| 2 | IL_8GLP_318 | 8GLP | 0.2413 | 2 | UAA/GAN related (H) | L5 | LSU rRNA | C | 4662 | G | 4663 | A | 4664 | A | 4665 | G | 4666 | * | U | 4632 | G | 4633 | A | 4635 | OMG | 4637 | cWW | cWW | ||
| 3 | IL_9E6Q_097 | 9E6Q | 0.1019 | 1 | UAA/GAN variation (H) | 1 | LSU rRNA | G | 2831 | G | 2832 | C | 2833 | A | 2834 | G | 2835 | * | U | 2802 | G | 2803 | A | 2805 | C | 2806 | cWW | tHS | cWW | |
| 4 | IL_4V9F_094 | 4V9F | 0.0000 | 1 | UAA/GAN variation | 0 | LSU rRNA | G | 2754 | G | 2755 | U | 2756 | A | 2757 | G | 2758 | * | U | 2724 | G | 2725 | A | 2727 | C | 2728 | cWW | tHS | cWW | |
| 5 | IL_9DFE_059 | 9DFE | 0.2416 | 0 | UAA/GAN variation | 1A | LSU rRNA | A | 1469 | G | 1470 | A | 1471 | A | 1472 | G | 1473 | * | U | 1518 | G | 1519 | G | 1520 | U | 1523 | cWW | tHS | cWW | |
| 6 | IL_9E6Q_021 | 9E6Q | 0.2985 | 0 | 1 | LSU rRNA | C | 588 | G | 589 | A | 590 | A | 591 | G | 592 | * | C | 616 | G | 617 | A | 618 | G | 619 | cWW | tSH | tHS | cWW | |
| 7 | IL_8B0X_122 | 8B0X | 0.2547 | 0 | UAA/GAN variation (H) | a | LSU rRNA | U | 1544 | G | 1545 | A | 1546 | A | 1547 | G | 1548 | * | C | 1528 | G | 1529 | A | 1530 | G | 1531 | cWW | tSH | tHS | cWW |
| 8 | IL_8XZL_001 | 8XZL | 0.2745 | 0 | UAA/GAN variation | A | THF-II riboswitch (folE RNA) | U | 6 | G | 7 | U | 8 | A | 9 | C | 10 | * | G | 45 | G | 46 | A | 47 | G | 48 | cWW | tSH | tHS | cWW |
| 9 | IL_8P9A_251 | 8P9A | 0.3045 | 0 | UAA/GAN variation | AR | LSU rRNA | U | 741 | G | 742 | C | 743 | A | 744 | C | 745 | * | G | 725 | G | 726 | G | 727 | G | 728 | cWW | ntSH | tHS | cWW |
| 10 | IL_8OI5_025 | 8OI5 | 0.3298 | 0 | 1 | LSU rRNA | U | 737 | G | 738 | C | 739 | A | 740 | C | 741 | * | G | 723 | G | 724 | G | 725 | G | 726 | cWW | ntSH | tHS | cWW | |
| 11 | IL_9AXU_022 | 9AXU | 0.3221 | 0 | 2 | LSU rRNA | U | 769 | G | 770 | C | 771 | A | 772 | C | 773 | * | G | 752 | G | 753 | A | 754 | A | 755 | cWW | tSH | tHS | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| UGCAC*GGGG | 2 |
| CCAAG*UGCAG | 1 |
| CGAAG*UGUA(PSU)(OMG) | 1 |
| GGCAG*UGUAC | 1 |
| GGUAG*UGUAC | 1 |
| AGAAG*UGGU | 1 |
| CGAAG*CGAG | 1 |
| UGAAG*CGAG | 1 |
| UGUAC*GGAG | 1 |
| UGCAC*GGAA | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GAA*GA | 2 |
| GCA*GG | 2 |
| CAA*GCA | 1 |
| GAA*GUA(PSU) | 1 |
| GCA*GUA | 1 |
| GUA*GUA | 1 |
| GAA*GG | 1 |
| GUA*GA | 1 |
| GCA*GA | 1 |
- Annotations
-
- UAA/GAN variation (5)
- (3)
- UAA/GAN variation (H) (2)
- UAA/GAN related (H) (1)
- Basepair signature
- cWW-tSH-L-tHS-cWW
- Heat map statistics
- Min 0.07 | Avg 0.29 | Max 0.49
Coloring options: