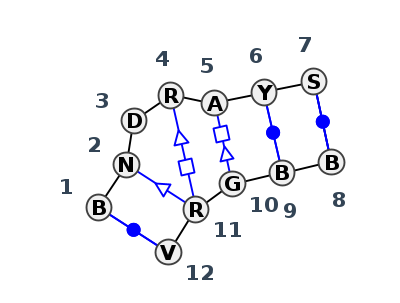
Motif IL_69229.1 Version IL_69229.1 of this group appears in releases 3.88 to 3.88
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | break | 8 | 9 | 10 | 11 | 12 | 1-12 | 2-11 | 4-11 | 5-10 | 6-9 | 7-8 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_6DVK_002 | 6DVK | 0.3696 | 2 | Kink-turn | H | RNA (95-MER) | C | 68 | G | 69 | U | 70 | G | 73 | A | 74 | C | 75 | G | 76 | * | C | 18 | C | 19 | G | 20 | A | 21 | G | 22 | cWW | tSH | tHS | ncBW | cWW | |
| 2 | IL_3Q3Z_002 | 3Q3Z | 0.1690 | 1 | Kink-turn | V | Cyclic di-GMP-II riboswitch | C | 16 | A | 17 | A | 18 | G | 20 | A | 21 | U | 22 | G | 23 | * | U | 54 | U | 55 | G | 56 | A | 57 | G | 58 | cWW | ntsS | tSH | tHS | cWw | cWW |
| 3 | IL_5GIP_001 | 5GIP | 0.0816 | 1 | Kink-turn | H+G | C/D RNA | U | 8 | U | 9 | G | 10 | G | 12 | A | 13 | U | 14 | G | 15 | * | C | 30 | U | 31 | G | 32 | A | 33 | A | 34 | cWW | ntsS | tSH | tHS | cWw | cWW |
| 4 | IL_5GIP_003 | 5GIP | 0.0867 | 1 | Kink-turn | G+H | C/D RNA | U | 8 | U | 9 | G | 10 | G | 12 | A | 13 | U | 14 | G | 15 | * | C | 30 | U | 31 | G | 32 | A | 33 | A | 34 | cWW | ntsS | tSH | tHS | cWw | cWW |
| 5 | IL_3NVI_002 | 3NVI | 0.0000 | 1 | Kink-turn | F | RNA (24-mer) | G | 15 | C | 16 | G | 17 | G | 19 | A | 20 | U | 21 | G | 22 | * | C | 4 | U | 5 | G | 6 | A | 7 | C | 8 | cWW | tsS | tSH | tHS | cWw | cWW |
| 6 | IL_3PLA_001 | 3PLA | 0.0739 | 1 | Kink-turn | G+H | C/D guide RNA | U | 8 | U | 9 | G | 10 | G | 12 | A | 13 | U | 14 | G | 15 | * | C | 29 | U | 30 | G | 31 | A | 32 | A | 33 | cWW | ntsS | tSH | tHS | cWw | cWW |
| 7 | IL_3NMU_004 | 3NMU | 0.0877 | 1 | Kink-turn | E | RNA (34-MER) | G | 24 | C | 25 | G | 26 | G | 28 | A | 29 | U | 30 | G | 31 | * | C | 13 | U | 14 | G | 15 | A | 16 | C | 17 | cWW | tSH | tHS | cWw | cWW | |
| 8 | IL_1RLG_004 | 1RLG | 0.1037 | 1 | Kink-turn | D | 25-MER | G | 15 | C | 16 | G | 17 | G | 19 | A | 20 | 5BU | 21 | G | 22 | * | C | 4 | 5BU | 5 | G | 6 | A | 7 | C | 8 | cWW | tsS | tSH | tHS | ncWw | cWW |
| 9 | IL_5G4U_005 | 5G4U | 0.1255 | 1 | Kink-turn | I+J | HMKT-7 | C | 3 | G | 4 | A | 5 | G | 7 | A | 8 | U | 9 | C | 10 | * | G | 13 | U | 14 | G | 15 | A | 16 | G | 17 | cWW | tsS | tSH | tHS | ncWw | cWW |
| 10 | IL_5G4U_006 | 5G4U | 0.1276 | 1 | Kink-turn | J+I | HMKT-7 | C | 3 | G | 4 | A | 5 | G | 7 | A | 8 | U | 9 | C | 10 | * | G | 13 | U | 14 | G | 15 | A | 16 | G | 17 | cWW | ntsS | tSH | tHS | ncWw | cWW |
| 11 | IL_5J7L_390 | 5J7L | 0.8593 | 10 | Kink-turn | DA | LSU rRNA | C | 2160 | C | 2161 | A | 2163 | A | 2171 | A | 2173 | C | 2174 | C | 2175 | * | G | 2123 | G | 2124 | G | 2125 | G | 2127 | G | 2128 | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| UUGUGAUG*CUGAA | 3 |
| GCGUGAUG*CUGAC | 2 |
| CGAAGAUC*GUGAG | 2 |
| CGUUUGACG*CCGAG | 1 |
| CAAUGAUG*UUGAG | 1 |
| GCGUGA(5BU)G*C(5BU)GAC | 1 |
| CCGACCUUGAAAUACC*GGGAGG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| UGUGAU*UGA | 3 |
| CGUGAU*UGA | 2 |
| GAAGAU*UGA | 2 |
| GUUUGAC*CGA | 1 |
| AAUGAU*UGA | 1 |
| CGUGA(5BU)*(5BU)GA | 1 |
| CGACCUUGAAAUAC*GGAG | 1 |
Release history
| Release | 3.88 |
|---|---|
| Date | 2024-09-11 |
| Status | New id, 1 parent |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- Kink-turn (11)
- Basepair signature
- cWW-tSS-tSH-L-tHS-cWW-cWW
- Heat map statistics
- Min 0.05 | Avg 0.28 | Max 0.97
Coloring options: