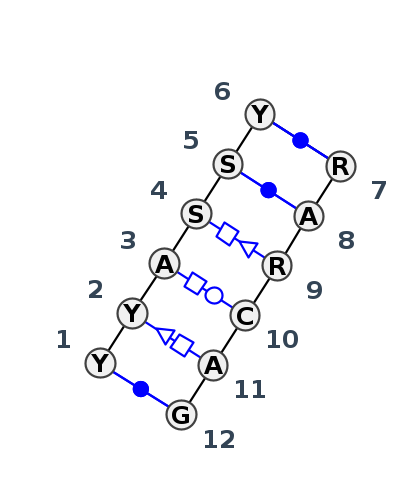
Motif IL_76308.5 Version IL_76308.5 of this group appears in releases 3.88 to 3.94
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | break | 7 | 8 | 9 | 10 | 11 | 12 | 1-12 | 2-10 | 2-11 | 3-10 | 4-9 | 5-8 | 6-7 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_6DVK_004 | 6DVK | 0.2524 | 0 | tSH-tWH-tHS-cWW | H | RNA (95-MER) | C | 52 | U | 53 | A | 54 | C | 55 | C | 56 | C | 57 | * | G | 38 | A | 39 | A | 40 | C | 41 | A | 42 | G | 43 | cWW | tSH | tHW | ncBW | cWW | ||
| 2 | IL_1MFQ_007 | 1MFQ | 0.1336 | 0 | tSH-tWH-tHS-cWW | A | SRP RNA | C | 190 | C | 191 | A | 192 | G | 193 | G | 194 | U | 195 | * | G | 204 | A | 205 | G | 206 | C | 207 | A | 208 | G | 209 | cWW | tWH | ntSH | tHW | tHS | cWW | cWW |
| 3 | IL_1LNT_002 | 1LNT | 0.0000 | 0 | tSH-tWH-tHS-cWW | A+B | RNA (12-mer) | U | 4 | C | 5 | A | 6 | G | 7 | G | 8 | U | 9 | * | A | 16 | A | 17 | G | 18 | C | 19 | A | 20 | G | 21 | cWW | tSH | tHW | tHS | cWW | cWW | |
| 4 | IL_3LQX_002 | 3LQX | 0.1491 | 0 | tSH-tWH-tHS-cWW | B | SRP RNA | U | 146 | C | 147 | A | 148 | G | 149 | G | 150 | U | 151 | * | A | 160 | A | 161 | G | 162 | C | 163 | A | 164 | G | 165 | cWW | tSH | tHW | tHS | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| UCAGGU*AAGCAG | 2 |
| CUACCC*GAACAG | 1 |
| CCAGGU*GAGCAG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| CAGG*AGCA | 3 |
| UACC*AACA | 1 |
Release history
| Release | 3.88 | 3.89 | 3.90 | 3.91 | 3.92 | 3.93 | 3.94 |
|---|---|---|---|---|---|---|---|
| Date | 2024-09-11 | 2024-10-09 | 2024-11-06 | 2024-12-04 | 2025-01-01 | 2025-01-29 | 2025-02-26 |
| Status | Updated, 1 parent | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- tSH-tWH-tHS-cWW (4)
- Basepair signature
- cWW-tSH-tHW-tHS-cWW-cWW
- Heat map statistics
- Min 0.13 | Avg 0.15 | Max 0.29
Coloring options: