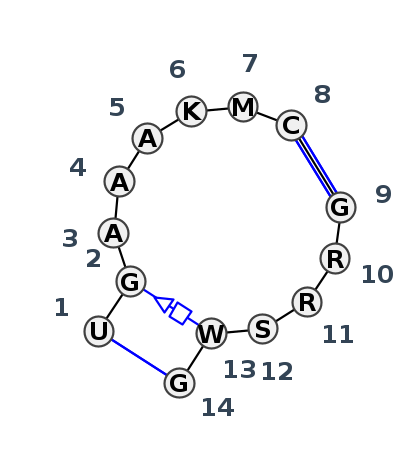
Motif IL_78349.2 Version IL_78349.2 of this group appears in releases 3.88 to 3.95
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | break | 9 | 10 | 11 | 12 | 13 | 14 | 1-14 | 2-13 | 7-10 | 7-11 | 8-9 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_5J7L_030 | 5J7L | 0.4951 | 0 | AA | SSU rRNA | U | 605 | G | 606 | A | 607 | A | 608 | A | 609 | U | 610 | C | 611 | C | 612 | * | G | 628 | A | 629 | A | 630 | C | 631 | U | 632 | G | 633 | cWW | cWWa | cWW | ||
| 2 | IL_4LFB_026 | 4LFB | 0.0000 | 0 | A | SSU rRNA | U | 605 | G | 606 | A | 607 | A | 608 | A | 609 | G | 610 | A | 611 | C | 612 | * | G | 628 | G | 629 | G | 630 | G | 631 | A | 632 | G | 633 | cWW | tSH | cWW | ||
| 3 | IL_6CZR_408 | 6CZR | 0.2637 | 0 | 1a | SSU rRNA | U | 589 | G | 590 | A | 591 | A | 592 | A | 593 | G | 594 | A | 595 | C | 596 | * | G | 612 | G | 613 | G | 614 | G | 615 | A | 616 | G | 617 | ncWW | tSH | ncWW | ncWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| UGAAAGAC*GGGGAG | 2 |
| UGAAAUCC*GAACUG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GAAAGA*GGGA | 2 |
| GAAAUC*AACU | 1 |
Release history
| Release | 3.88 | 3.89 | 3.90 | 3.91 | 3.92 | 3.93 | 3.94 | 3.95 |
|---|---|---|---|---|---|---|---|---|
| Date | 2024-09-11 | 2024-10-09 | 2024-11-06 | 2024-12-04 | 2025-01-01 | 2025-01-29 | 2025-02-26 | 2025-03-26 |
| Status | Updated, 1 parent | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (3)
- Basepair signature
- cWW-tSH-L-R-L-R-L-R-L-cWW-L
- Heat map statistics
- Min 0.26 | Avg 0.29 | Max 0.56
Coloring options: