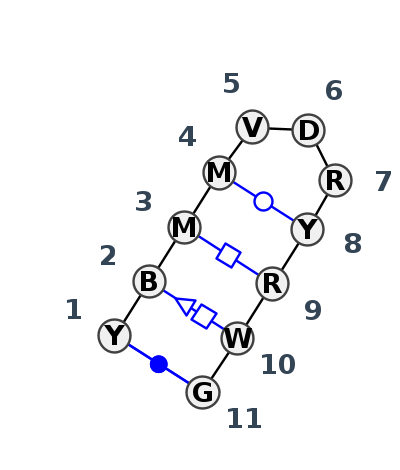
Motif HL_19870.9 Version HL_19870.9 of this group appears in releases 3.89 to 3.90
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 1-11 | 2-10 | 3-9 | 4-8 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HL_4V9F_059 | 4V9F | 0.9662 | 0 | 0 | LSU rRNA | U | 2389 | U | 2390 | C | 2391 | C | 2392 | C | 2393 | A | 2394 | A | 2395 | C | 2396 | G | 2397 | A | 2398 | G | 2399 | cWW | tSH | tHH | |
| 2 | HL_8C3A_056 | 8C3A | 0.1267 | 2 | 1 | LSU rRNA | U | 2605 | U | 2606 | A | 2607 | A | 2608 | C | 2610 | G | 2611 | A | 2612 | U | 2613 | A | 2614 | A | 2615 | G | 2617 | cWW | ntSH | tHH | tWW |
| 3 | HL_5TBW_055 | 5TBW | 0.1203 | 2 | 1 | LSU rRNA | U | 2633 | U | 2634 | A | 2635 | A | 2636 | C | 2638 | G | 2639 | A | 2640 | U | 2641 | A | 2642 | A | 2643 | G | 2645 | cWW | ntSH | tHH | tWW |
| 4 | HL_4V9F_056 | 4V9F | 0.1195 | 2 | 0 | LSU rRNA | C | 2298 | G | 2299 | A | 2300 | A | 2301 | A | 2303 | G | 2304 | A | 2305 | U | 2306 | A | 2307 | U | 2308 | G | 2310 | cWW | tSH | tHH | tWW |
| 5 | HL_4WF9_050 | 4WF9 | 0.0000 | 2 | X | LSU rRNA | C | 2291 | U | 2292 | A | 2293 | A | 2294 | A | 2296 | G | 2297 | G | 2298 | U | 2299 | A | 2300 | A | 2301 | G | 2303 | cWW | ntSH | tHH | tWW |
| 6 | HL_5J7L_187 | 5J7L | 0.0967 | 2 | DA | LSU rRNA | C | 2264 | U | 2265 | A | 2266 | A | 2267 | G | 2269 | A | 2270 | G | 2271 | U | 2272 | A | 2273 | A | 2274 | G | 2276 | cWW | ntSH | tHH | tWW |
| 7 | HL_8VTW_053 | 8VTW | 0.0841 | 2 | 1A | LSU rRNA | C | 2264 | U | 2265 | A | 2266 | A | 2267 | A | 2269 | G | 2270 | G | 2271 | U | 2272 | A | 2273 | A | 2274 | G | 2276 | cWW | ntSH | tHH | tWW |
| 8 | HL_7A0S_053 | 7A0S | 0.1378 | 2 | X | LSU rRNA | C | 2243 | C | 2244 | A | 2245 | A | 2246 | A | 2248 | U | 2249 | G | 2250 | U | 2251 | A | 2252 | A | 2253 | G | 2255 | cWW | ntSH | ntHH | tWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| UUAAACGAUAACG | 2 |
| CUAAAAGGUAACG | 2 |
| UUCCCAACGAG | 1 |
| CGAAAAGAUAUCG | 1 |
| CUAAAGAGUAACG | 1 |
| CCAAAAUGUAACG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| UAAACGAUAAC | 2 |
| UAAAAGGUAAC | 2 |
| UCCCAACGA | 1 |
| GAAAAGAUAUC | 1 |
| UAAAGAGUAAC | 1 |
| CAAAAUGUAAC | 1 |
- Annotations
-
- (8)
- Basepair signature
- cWW-tHH-cWW-tHS-F-F-tWW
- Heat map statistics
- Min 0.08 | Avg 0.30 | Max 0.98
Coloring options: