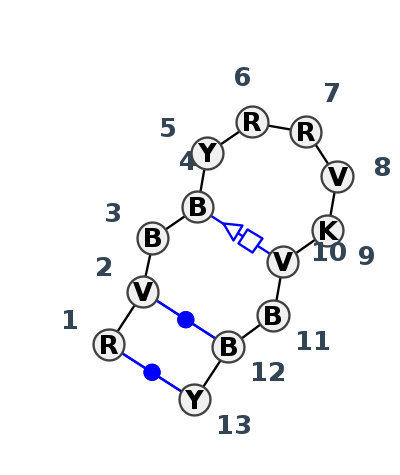
Motif HL_68749.1 Version HL_68749.1 of this group appears in releases 4.0 to 4.6
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 1-13 | 2-12 | 3-11 | 4-9 | 4-10 | 5-8 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HL_8GLP_057 | 8GLP | 0.6736 | 1 | L5 | LSU rRNA | A | 4098 | G | 4099 | C | 4100 | C | 4101 | C | 4103 | G | 4104 | A | 4105 | G | 4106 | G | 4107 | G | 4108 | G | 4109 | C | 4110 | U | 4111 | cWW | ncWW | ncWH | |||
| 2 | HL_8GLP_097 | 8GLP | 0.2623 | 0 | S2 | SSU rRNA | A | 569 | C | 570 | U | 571 | PSU | 572 | PSU | 573 | A | 574 | A | 575 | A2M | 576 | U | 577 | C | 578 | C | 579 | U | 580 | U | 581 | cWW | cWW | tSH | |||
| 3 | HL_8CRE_204 | 8CRE | 0.0937 | 0 | CM | SSU rRNA | A | 518 | A | 519 | U | 520 | G | 521 | U | 522 | A | 523 | A | 524 | A | 525 | U | 526 | A | 527 | C | 528 | C | 529 | U | 530 | cWW | cWWa | tSH | |||
| 4 | HL_8P9A_203 | 8P9A | 0.0000 | 0 | sR | SSU rRNA | A | 520 | A | 521 | U | 522 | G | 523 | U | 524 | A | 525 | A | 526 | A | 527 | U | 528 | A | 529 | C | 530 | C | 531 | U | 532 | cWW | ncWWa | ntSH | |||
| 5 | HL_4WFL_001 | 4WFL | 0.2729 | 0 | A | LSRP RNA | G | 19 | A | 20 | G | 21 | G | 22 | U | 23 | A | 24 | G | 25 | C | 26 | G | 27 | G | 28 | U | 29 | G | 30 | C | 31 | cWW | cWW | tSH | ntSHa |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| AAUGUAAAUACCU | 2 |
| AGCCCCGAGGGGCU | 1 |
| ACUAAUCCUU | 1 |
| GAGGUAGCGGUGC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| AUGUAAAUACC | 2 |
| GCCCCGAGGGGC | 1 |
| CU(PSU)(PSU)AA(A2M)UCCU | 1 |
| AGGUAGCGGUG | 1 |
Release history
| Release | 4.0 | 4.1 | 4.2 | 4.3 | 4.4 | 4.5 | 4.6 |
|---|---|---|---|---|---|---|---|
| Date | 2025-08-13 | 2025-09-10 | 2025-10-08 | 2025-11-05 | 2025-12-03 | 2025-12-31 | 2026-01-28 |
| Status | New id, 1 parent | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (5)
- Basepair signature
- cWW-cWW-F-F-tSH-F-F-F-F-F
- Heat map statistics
- Min 0.09 | Avg 0.34 | Max 0.70
Coloring options: