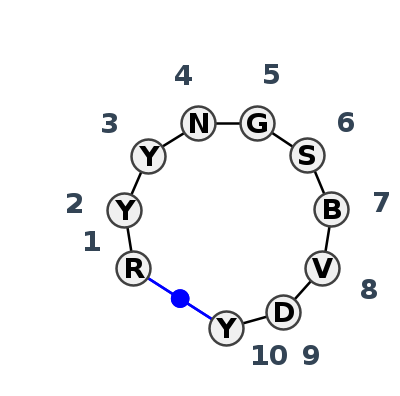
Motif HL_92697.1 Version HL_92697.1 of this group appears in releases 4.0 to 4.6
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 1-10 | 2-9 | 4-6 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HL_5X2G_002 | 5X2G | 0.8043 | 1 | B | sgRNA | G | 54 | U | 55 | U | 56 | U | 57 | G | 58 | C | 59 | G | 60 | G | 61 | G | 62 | C | 64 | cWW | ncSH | |
| 2 | HL_7MKY_001 | 7MKY | 0.6407 | 2 | A | RNA (66-MER) | G | 9 | U | 10 | C | 12||A | A | 13 | G | 14 | C | 15 | C | 16 | C | 17 | G | 18 | C | 20 | cWW | cBW | |
| 3 | HL_4L47_036 | 4L47 | 0.3298 | 0 | QY | messenger RNA | G | 31 | U | 32 | U | 33 | C | 34 | G | 35 | G | 36 | 1MG | 37 | G | 37|||A | A | 38 | C | 39 | cWW | ||
| 4 | HL_3B31_001 | 3B31 | 0.0000 | 0 | A | RNA (29-MER) | A | 6184 | U | 6185 | U | 6186 | A | 6187 | G | 6188 | G | 6189 | U | 6190 | A | 6191 | G | 6192 | U | 6193 | cWW | ||
| 5 | HL_6XKO_003 | 6XKO | 0.5891 | 0 | A | PreQ1-III riboswitch | G | 63 | C | 64 | U | 65 | G | 66 | G | 67 | C | 68 | G | 69 | G | 70 | U | 71 | C | 72 | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GUUUGCGGGAC | 1 |
| GUGCAGCCCGUC | 1 |
| GUUCGGGAC | 1 |
| AUUAGGUAGU | 1 |
| GCUGGCGGUC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| UUUGCGGGA | 1 |
| UGCAGCCCGU | 1 |
| UUCGG(1MG)GA | 1 |
| UUAGGUAG | 1 |
| CUGGCGGU | 1 |
Release history
| Release | 4.0 | 4.1 | 4.2 | 4.3 | 4.4 | 4.5 | 4.6 |
|---|---|---|---|---|---|---|---|
| Date | 2025-08-13 | 2025-09-10 | 2025-10-08 | 2025-11-05 | 2025-12-03 | 2025-12-31 | 2026-01-28 |
| Status | New id, 2 parents | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (5)
- Basepair signature
- cWW-F-F-F-F-F-F-F-F
- Heat map statistics
- Min 0.33 | Avg 0.56 | Max 0.99
Coloring options: