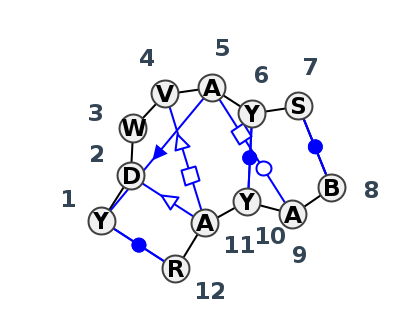
Motif IL_46174.4 Version IL_46174.4 of this group appears in releases 4.0 to 4.6
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | break | 8 | 9 | 10 | 11 | 12 | 1-5 | 1-12 | 2-11 | 4-11 | 5-9 | 6-10 | 7-8 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_4LFB_011 | 4LFB | 0.1517 | 2 | Kink-turn with non-sequential stacking | A | SSU rRNA | C | 277 | G | 278 | A | 279 | G | 281 | A | 282 | C | 283 | G | 284 | * | C | 242 | A | 243 | C | 245 | A | 246 | G | 247 | ncsS | cWW | ntsS | tSH | tHW | cwW | cWW |
| 2 | IL_8B0X_013 | 8B0X | 0.1252 | 2 | Kink-turn with non-sequential stacking (H) | A | SSU rRNA | C | 277 | G | 278 | A | 279 | G | 281 | A | 282 | U | 283 | C | 284 | * | G | 242 | A | 243 | U | 245 | A | 246 | G | 247 | ncsS | cWW | ntsS | tSH | tHW | cWw | cWW |
| 3 | IL_8GLP_210 | 8GLP | 0.0535 | 2 | Kink-turn with non-sequential stacking (H) | S2 | SSU rRNA | U | 396 | G | 397 | A | 398 | C | 400 | A | 401 | C | 402 | G | 403 | * | U | 359 | A | 360 | C | 362 | A | 363 | A | 364 | ncsS | cWW | ntsS | tHW | cwW | cWW | |
| 4 | IL_8CRE_408 | 8CRE | 0.0000 | 2 | Kink-turn with non-sequential stacking | CM | SSU rRNA | U | 346 | U | 347 | U | 348 | A | 350 | A | 351 | C | 352 | G | 353 | * | U | 309 | A | 310 | C | 312 | A | 313 | A | 314 | ncsS | cWW | tsS | ntSH | tHW | cwW | cWW |
| 5 | IL_8P9A_391 | 8P9A | 0.0528 | 2 | Kink-turn with non-sequential stacking | sR | SSU rRNA | U | 348 | U | 349 | U | 350 | A | 352 | A | 353 | C | 354 | G | 355 | * | U | 311 | A | 312 | C | 314 | A | 315 | A | 316 | csS | cWW | tsS | ntSH | tHW | cwW | cWW |
| 6 | IL_3RW6_002 | 3RW6 | 0.2768 | 4 | Kink-turn with non-sequential stacking | H | constitutive transport element(CTE)of Mason-Pfizer monkey virus RNA | C | 43 | A | 44 | A | 45 | G | 47 | A | 48 | C | 49 | G | 50 | * | U | 11 | A | 12 | C | 16 | A | 17 | G | 18 | ncsS | cWW | tsS | tSH | tHW | cwW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| UUUCAACG*UAUCAA | 2 |
| CGACGACG*CAUCAG | 1 |
| CGACGAUC*GAUUAG | 1 |
| UGACCACG*UAUCAA | 1 |
| CAAUGACG*UAAGACAG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| UUCAAC*AUCA | 2 |
| GACGAC*AUCA | 1 |
| GACGAU*AUUA | 1 |
| GACCAC*AUCA | 1 |
| AAUGAC*AAGACA | 1 |
Release history
| Release | 4.0 | 4.1 | 4.2 | 4.3 | 4.4 | 4.5 | 4.6 |
|---|---|---|---|---|---|---|---|
| Date | 2025-08-13 | 2025-09-10 | 2025-10-08 | 2025-11-05 | 2025-12-03 | 2025-12-31 | 2026-01-28 |
| Status | Updated, 1 parent | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- Kink-turn with non-sequential stacking (4)
- Kink-turn with non-sequential stacking (H) (2)
- Basepair signature
- cWW-cSS-tSS-tSH-L-cWW-tHW-cWW
- Heat map statistics
- Min 0.05 | Avg 0.14 | Max 0.31
Coloring options: