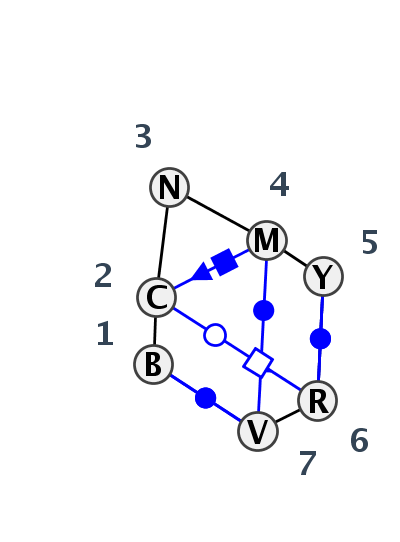
Motif IL_63596.6 Version IL_63596.6 of this group appears in releases 3.80 to 3.86
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | break | 6 | 7 | 1-3 | 1-7 | 2-4 | 2-6 | 3-7 | 4-7 | 5-6 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_7RQB_051 | 7RQB | 0.2212 | 1 | C-loop | 1A | LSU rRNA | G | 1319 | C | 1320 | A | 1321 | A | 1322 | U | 1323 | * | A | 1331 | C | 1333 | cWW | ncSH | tWH | cWS | cWW | ||
| 2 | IL_5J7L_291 | 5J7L | 0.2173 | 1 | C-loop | DA | LSU rRNA | C | 1319 | C | 1320 | A | 1321 | A | 1322 | C | 1323 | * | G | 1331 | G | 1333 | cWW | ncSH | tWH | cWS | cWW | ||
| 3 | IL_4V9F_093 | 4V9F | 0.0000 | 1 | C-loop | 0 | LSU rRNA | C | 2717 | C | 2718 | A | 2719 | C | 2720 | U | 2721 | * | A | 2761 | G | 2763 | cWW | cSH | tWH | cWS | cWW | ||
| 4 | IL_4V9F_051 | 4V9F | 0.1314 | 1 | C-loop | 0 | LSU rRNA | G | 1425 | C | 1426 | A | 1427 | C | 1428 | U | 1429 | * | A | 1437 | C | 1439 | cWW | cSH | tWH | cWS | cWW | ||
| 5 | IL_5J7L_272 | 5J7L | 0.1478 | 2 | C-loop with bulged stacked A's | DA | LSU rRNA | G | 864 | C | 865 | A | 866 | C | 867 | U | 868 | * | A | 909 | C | 912 | cWW | cSH | tWH | cWS | cWW | ||
| 6 | IL_7RQB_104 | 7RQB | 0.1326 | 1 | C-loop | 1A | LSU rRNA | C | 2680 | C | 2681 | U | 2682 | C | 2683 | U | 2684 | * | A | 2725 | G | 2727 | cWW | cSH | tWH | tHS | cWS | cWW | |
| 7 | IL_4WF9_101 | 4WF9 | 0.1274 | 1 | C-loop | X | LSU rRNA | C | 2707 | C | 2708 | U | 2709 | C | 2710 | U | 2711 | * | A | 2752 | G | 2754 | cWW | ncSH | tWH | ntHS | cWS | cWW | |
| 8 | IL_7A0S_095 | 7A0S | 0.1644 | 1 | C-loop | X | LSU rRNA | C | 2659 | C | 2660 | G | 2661 | C | 2662 | U | 2663 | * | A | 2705 | G | 2707 | cWW | ncSH | tWH | cWS | cWW | ||
| 9 | IL_5J7L_347 | 5J7L | 0.1749 | 1 | C-loop | DA | LSU rRNA | U | 2680 | C | 2681 | A | 2682 | C | 2683 | U | 2684 | * | A | 2725 | A | 2727 | ncSH | cWW | cSH | tWH | cWS | cWW | |
| 10 | IL_4V88_443 | 4V88 | 0.2541 | 2 | C-loop | A6 | SSU rRNA | U | 1617 | C | 1618 | C | 1619 | C | 1620 | U | 1621 | * | A | 1157 | A | 1160 | ncSH | cWW | ncSH | tWH | cWS | cWW | |
| 11 | IL_6JDV_004 | 6JDV | 0.2575 | 1 | C-loop | B | CRISPR RNA direct repeat element | G | 105 | C | 106 | U | 107 | C | 108 | U | 109 | * | A | 138 | C | 140 | ncSH | cWW | tWH | cWS | cWW | ||
| 12 | IL_8HNT_002 | 8HNT | 0.2088 | 1 | C-loop | B | sgRNA | G | 87 | C | 88 | U | 89 | C | 90 | U | 91 | * | A | 120 | C | 122 | ncSH | cWW | ncSH | tWH | cWS | cWW | |
| 13 | IL_5B2P_003 | 5B2P | 0.2083 | 1 | C-loop | B | Guide RNA | C | 76 | C | 77 | U | 78 | C | 79 | U | 80 | * | A | 88 | G | 90 | cWW | cSH | tWH | tHS | cWS | cWW | |
| 14 | IL_4JRC_003 | 4JRC | 0.2199 | 1 | C-loop | B | Distal Stem I region of the glyQS T box leader RNA | C | 29 | C | 30 | U | 31 | C | 32 | U | 33 | * | A | 73 | G | 75 | cWW | cSH | tWH | tHS | cWS | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CCUCU*AUG | 2 |
| GCUCU*AUC | 2 |
| CCUCU*ACG | 2 |
| GCAAU*AGC | 1 |
| CCAAC*GGG | 1 |
| CCACU*ACG | 1 |
| GCACU*AGC | 1 |
| GCACU*AAAC | 1 |
| CCGCU*AUG | 1 |
| UCACU*AAA | 1 |
| UCCCU*ACCA | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| CUC*U | 4 |
| CAA*G | 2 |
| CUC*C | 2 |
| CAC*C | 1 |
| CAC*G | 1 |
| CAC*AA | 1 |
| CGC*U | 1 |
| CAC*A | 1 |
| CCC*CC | 1 |
Release history
| Release | 3.80 | 3.81 | 3.82 | 3.83 | 3.84 | 3.85 | 3.86 |
|---|---|---|---|---|---|---|---|
| Date | 2024-01-31 | 2024-02-28 | 2024-03-27 | 2024-04-24 | 2024-05-22 | 2024-06-19 | 2024-07-17 |
| Status | Updated, 1 parent | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- C-loop (13)
- C-loop with bulged stacked A's (1)
- Basepair signature
- cWW-cWS-cSH-tWH-cWW-L
- Heat map statistics
- Min 0.11 | Avg 0.18 | Max 0.33
Coloring options: