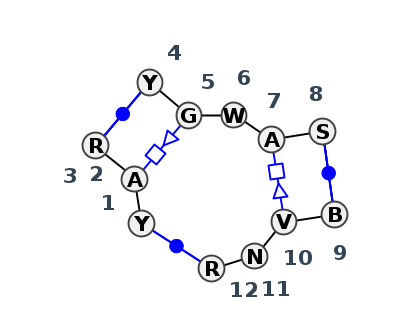
Motif J3_69816.1 Version J3_69816.1 of this group appears in releases 4.0 to 4.0
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | break | 4 | 5 | 6 | 7 | 8 | break | 9 | 10 | 11 | 12 | 1-12 | 2-5 | 3-4 | 7-10 | 8-9 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | J3_8GLP_003 | 8GLP | 0.2666 | 0 | L5 | LSU rRNA | C | 1546 | A | 1547 | G | 1548 | * | C | 1580 | G | 1581 | PSU | 1582 | A | 1583 | G | 1584 | * | U | 1609 | C | 1610 | C | 1611 | G | 1612 | cWW | tHS | cWW | tHS | cWW |
| 2 | J3_9AXU_010 | 9AXU | 0.2120 | 0 | 2 | LSU rRNA | U | 861 | A | 862 | G | 863 | * | C | 895 | G | 896 | U | 897 | A | 898 | G | 899 | * | U | 924 | C | 925 | A | 926 | A | 927 | cWW | tHS | cWW | tHS | cWW |
| 3 | J3_8OI5_010 | 8OI5 | 0.1765 | 0 | 1 | LSU rRNA | U | 825 | A | 826 | G | 827 | * | C | 859 | G | 860 | U | 861 | A | 862 | G | 863 | * | U | 888 | C | 889 | G | 890 | A | 891 | cWW | tHS | cWW | tHS | cWW |
| 4 | J3_9E6Q_008 | 9E6Q | 0.0000 | 0 | 1 | LSU rRNA | C | 815 | A | 816 | G | 817 | * | C | 849 | G | 850 | A | 851 | A | 852 | G | 853 | * | C | 878 | A | 879 | U | 880 | G | 881 | cWW | tHS | cWW | tHS | cWW |
| 5 | J3_9DFE_008 | 9DFE | 0.0907 | 0 | 1A | LSU rRNA | C | 698 | A | 699 | G | 700 | * | C | 732 | G | 733 | A | 734 | A | 735 | C | 736 | * | G | 760 | A | 761 | U | 762 | G | 763 | cWW | tHS | cWW | tHS | cWW |
| 6 | J3_8B0X_017 | 8B0X | 0.0875 | 0 | a | LSU rRNA | C | 700 | A | 701 | G | 702 | * | C | 734 | G | 735 | A | 736 | A | 737 | C | 738 | * | G | 762 | A | 763 | U | 764 | G | 765 | cWW | tHS | cWW | tHS | cWW |
| 7 | J3_4WF9_003 | 4WF9 | 0.0858 | 0 | X | LSU rRNA | C | 743 | A | 744 | G | 745 | * | C | 777 | G | 778 | A | 779 | A | 780 | C | 781 | * | G | 805 | A | 806 | U | 807 | G | 808 | cWW | tHS | cWW | tHS | cWW |
| 8 | J3_7A0S_005 | 7A0S | 0.1152 | 0 | X | LSU rRNA | C | 711 | A | 712 | G | 713 | * | C | 745 | G | 746 | A | 747 | A | 748 | C | 749 | * | G | 773 | A | 774 | U | 775 | G | 776 | cWW | tHS | cWW | ntHS | cWW |
| 9 | J3_4V9F_003 | 4V9F | 0.2130 | 1 | 0 | LSU rRNA | C | 789 | A | 790 | A | 791 | * | U | 823 | G | 824 | U | 826 | A | 827 | G | 828 | * | C | 853 | G | 854 | U | 855 | G | 856 | cWW | tHS | cWW | tHS | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CAG*CGAAC*GAUG | 4 |
| CAG*CG(PSU)AG*UCCG | 1 |
| UAG*CGUAG*UCAA | 1 |
| UAG*CGUAG*UCGA | 1 |
| CAG*CGAAG*CAUG | 1 |
| CAA*UGUUAG*CGUG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| A*GAA*AU | 5 |
| A*G(PSU)A*CC | 1 |
| A*GUA*CA | 1 |
| A*GUA*CG | 1 |
| A*GUUA*GU | 1 |
Release history
| Release | 4.0 |
|---|---|
| Date | 2025-08-13 |
| Status | New id, 1 parent |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (9)
- Basepair signature
- cWW-tHS-F-cWW-tHS-cWW-F
- Heat map statistics
- Min 0.07 | Avg 0.17 | Max 0.31
Coloring options: