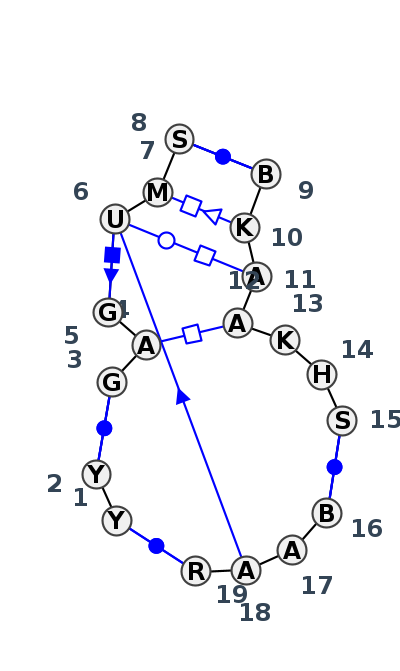
Motif J4_45801.2 Version J4_45801.2 of this group appears in releases 3.48 to 3.53
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | break | 3 | 4 | 5 | 6 | 7 | 8 | break | 9 | 10 | 11 | 12 | 13 | 14 | 15 | break | 16 | 17 | 18 | 19 | 1-19 | 2-3 | 4-12 | 5-6 | 6-11 | 6-18 | 7-10 | 8-9 | 11-17 | 12-15 | 12-17 | 12-18 | 15-16 | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | J4_4Y4O_014 | 4Y4O | 0.2283 | 1 | 2A | LSU rRNA | C | 268 | U | 269 | * | G | 370 | A | 371 | G | 372 | U | 373 | A | 374 | C | 375 | * | G | 399 | G | 400 | A | 401 | A | 402 | U | 403 | C | 404 | G | 406 | * | U | 421 | A | 422 | A | 423 | G | 424 | cWW | cWW | tHH | cSH | tWH | ncSs | tHS | cWW | ntWW | ntWW | cWW | ||
| 2 | J4_4V9F_001 | 4V9F | 0.2003 | 1 | 0 | LSU rRNA | C | 239 | C | 240 | * | G | 379 | A | 380 | G | 381 | U | 382 | A | 383 | G | 384 | * | C | 405 | G | 406 | A | 407 | A | 408 | U | 409 | A | 410 | C | 412 | * | G | 428 | A | 429 | A | 430 | G | 431 | cWW | cWW | tHH | cSH | tWH | cSs | tHS | cWW | ntWW | ntWW | cWW | ||
| 3 | J4_5J7L_018 | 5J7L | 0.2005 | 1 | DA | LSU rRNA | C | 268 | C | 269 | * | G | 370 | A | 371 | G | 372 | U | 373 | A | 374 | G | 375 | * | U | 399 | G | 400 | A | 401 | A | 402 | U | 403 | A | 404||A | G | 406 | * | C | 421 | A | 422 | A | 423 | G | 424 | cWW | cWW | tHH | cSH | tWH | cSs | tHS | cWW | ntWW | ntWW | ntWW | cWW | |
| 4 | J4_4WF9_001 | 4WF9 | 0.2061 | 1 | X | LSU rRNA | C | 271 | C | 272 | * | G | 416 | A | 417 | G | 418 | U | 419 | A | 420 | C | 421 | * | G | 445 | G | 446 | A | 447 | A | 448 | U | 449 | C | 450 | G | 452 | * | U | 467 | A | 468 | A | 469 | G | 470 | cWW | cWW | ntHH | cSH | ntWH | cSs | tHS | cWW | ntWW | ncsS | ntWW | ncWW | |
| 5 | J4_7A0S_001 | 7A0S | 0.0000 | 1 | X | LSU rRNA | C | 245 | C | 246 | * | G | 383 | A | 384 | G | 385 | U | 386 | A | 387 | G | 388 | * | U | 412 | G | 413 | A | 414 | A | 415 | U | 416 | C | 417 | G | 419 | * | C | 434 | A | 435 | A | 436 | G | 437 | cWW | cWW | ntHH | cSH | tWH | ntHS | cWW | ntWW | ntWW | cWW | |||
| 6 | J4_5TBW_001 | 5TBW | 0.4827 | 0 | 1 | LSU rRNA | U | 112 | C | 113 | * | G | 267 | A | 268 | G | 269 | U | 270 | C | 271 | G | 272 | * | C | 293 | U | 294 | A | 295 | A | 296 | G | 297 | U | 298 | G | 299 | * | U | 316 | A | 317 | A | 318 | A | 319 | cWW | cWW | tHH | cSH | tWH | cSs | tHS | cWW | ntWW | ntWW | ntWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CU*GAGUAC*GGAAUCUG*UAAG | 1 |
| CC*GAGUAG*CGAAUAAC*GAAG | 1 |
| CC*GAGUAG*UGAAUAUG*CAAG | 1 |
| CC*GAGUAC*GGAAUCUG*UAAG | 1 |
| CC*GAGUAG*UGAAUCCG*CAAG | 1 |
| UC*GAGUCG*CUAAGUG*UAAA | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| *AGUA*GAAUCU*AA | 2 |
| *AGUA*GAAUAA*AA | 1 |
| *AGUA*GAAUAU*AA | 1 |
| *AGUA*GAAUCC*AA | 1 |
| *AGUC*UAAGU*AA | 1 |
- Annotations
-
- (6)
- Basepair signature
- cWW-cWW-cSS-F-tHH-cWW-cSH-tWH-F-tHS-F-cWW
- Heat map statistics
- Min 0.16 | Avg 0.26 | Max 0.57
Coloring options: