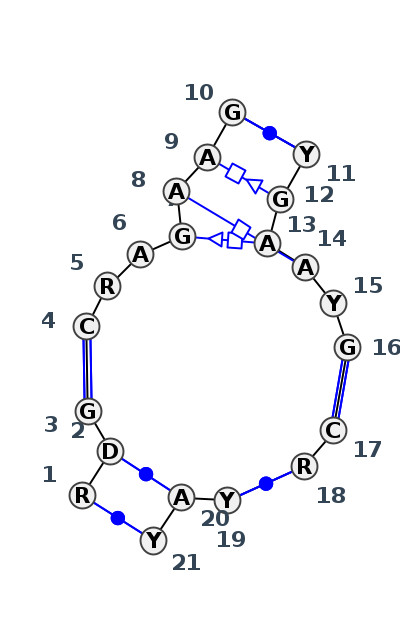
Motif J5_27223.1 Version J5_27223.1 of this group appears in releases 4.0 to 4.6
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | break | 4 | 5 | 6 | 7 | 8 | 9 | 10 | break | 11 | 12 | 13 | 14 | 15 | 16 | break | 17 | 18 | break | 19 | 20 | 21 | 1-21 | 2-20 | 3-4 | 3-8 | 4-8 | 5-6 | 6-15 | 7-13 | 7-15 | 8-14 | 9-12 | 10-11 | 16-17 | 18-19 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | J5_4WF9_001 | 4WF9 | 0.2288 | 1 | X | LSU rRNA | A | 178 | A | 179 | G | 180 | * | C | 47 | A | 48 | A | 49 | G | 51 | A | 52 | A | 53 | G | 54 | * | C | 115 | G | 116 | A | 117 | A | 118 | U | 119 | G | 120 | * | C | 129 | A | 130 | * | U | 149 | A | 150 | U | 151 | ncWW | ncwW | cWW | tsS | ncsS | tSH | ntHH | tHS | cWW | cWW | cWW | |||
| 2 | J5_9DFE_001 | 9DFE | 0.2472 | 1 | 1A | LSU rRNA | G | 176 | G | 177 | G | 178 | * | C | 47 | G | 48 | A | 49 | G | 51 | A | 52 | A | 53 | G | 54 | * | C | 116 | G | 117 | A | 118 | A | 119 | U | 120 | G | 121 | * | C | 130 | G | 131 | * | C | 148 | A | 149 | C | 150 | cWW | cWW | ncsS | ncSs | cHW | tSH | tHH | tHS | cWW | cWW | cWW | |||
| 3 | J5_8B0X_004 | 8B0X | 0.2075 | 1 | a | LSU rRNA | A | 176 | G | 177 | G | 178 | * | C | 47 | G | 48 | A | 49 | G | 51 | A | 52 | A | 53 | G | 54 | * | C | 116 | G | 117 | A | 118 | A | 119 | U | 120 | G | 121 | * | C | 130 | A | 131 | * | U | 148 | A | 149 | U | 150 | cWW | ncHW | cWW | ncsS | ncSs | cHW | tSH | tHH | tHS | cWW | cWW | cWW | ||
| 4 | J5_8GLP_001 | 8GLP | 0.0987 | 1 | L5+L8 | LSU rRNA + 5.8S rRNA | G | 19 | U | 20 | G | 21 | * | C | 35 | G | 36 | A | 37 | G | 39 | A | 40 | A | 41 | G | 42 | * | C | 101 | G | 102 | A | 103 | A | 104 | C | 105 | G | 106 | * | C | 113 | G | 114 | * | U | 136 | A | 137 | C | 138 | cWW | cWW | ncSs | tSH | ntWW | tHH | tHS | cWW | cWW | cWW | ||||
| 5 | J5_8P9A_010 | 8P9A | 0.0000 | 1 | AR+AT | LSU rRNA + 5.8S rRNA | A | 20 | G | 21 | G | 22 | * | C | 35 | G | 36 | A | 37 | G | 39 | A | 40 | A | 41 | G | 42 | * | U | 102 | G | 103 | A | 104 | A | 105 | C | 106 | G | 107 | * | C | 115 | G | 116 | * | C | 137 | A | 138 | U | 139 | cWW | cWW | cWW | tSH | tWW | tHH | tHS | cWW | cWW | cWW | ||||
| 6 | J5_8OI5_001 | 8OI5 | 0.0479 | 1 | 1+4 | LSU rRNA + 5.8S rRNA | A | 19 | G | 20 | G | 21 | * | C | 35 | G | 36 | A | 37 | G | 39 | A | 40 | A | 41 | G | 42 | * | U | 102 | G | 103 | A | 104 | A | 105 | C | 106 | G | 107 | * | C | 115 | G | 116 | * | C | 137 | A | 138 | U | 139 | cWW | cWW | cWW | ncsS | ncSs | tSH | ntWW | ntHH | tHS | cWW | cWW | cWW | ||
| 7 | J5_9AXU_001 | 9AXU | 0.0577 | 1 | 2+4 | LSU rRNA + 5.8S rRNA | A | 20 | G | 21 | G | 22 | * | C | 43 | G | 44 | A | 45 | G | 47 | A | 48 | A | 49 | G | 50 | * | U | 110 | G | 111 | A | 112 | A | 113 | C | 114 | G | 115 | * | C | 123 | G | 124 | * | C | 145 | A | 146 | U | 147 | cWW | cWW | cWW | ncSs | tSH | tWW | tHH | tHS | cWW | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| AGG*CGAUGAAG*UGAACG*CG*CAU | 3 |
| CAAUGAAG*CGAAUG*CA*UAU*AAG | 1 |
| CGAUGAAG*CGAAUG*CG*CAC*GGG | 1 |
| CGAUGAAG*CGAAUG*CA*UAU*AGG | 1 |
| GUG*CGAUGAAG*CGAACG*CG*UAC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| G*GAUGAA*GAAC**A | 3 |
| GAUGAA*GAAU**A*G | 2 |
| AAUGAA*GAAU**A*A | 1 |
| U*GAUGAA*GAAC**A | 1 |
Release history
| Release | 4.0 | 4.1 | 4.2 | 4.3 | 4.4 | 4.5 | 4.6 |
|---|---|---|---|---|---|---|---|
| Date | 2025-08-13 | 2025-09-10 | 2025-10-08 | 2025-11-05 | 2025-12-03 | 2025-12-31 | 2026-01-28 |
| Status | New id, 1 parent | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (7)
- Basepair signature
- cWW-cWW-cWW-cWW-F-cWW-F-tSH-F-tHH-tHS-cWW
- Heat map statistics
- Min 0.05 | Avg 0.15 | Max 0.26
Coloring options: