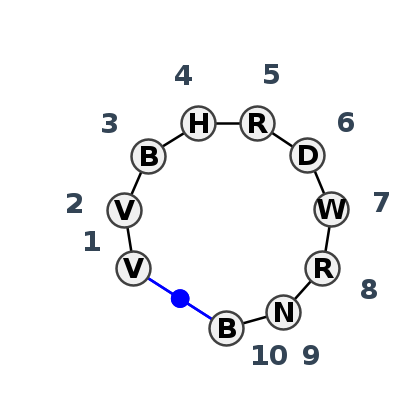
Motif HL_75293.5 Version HL_75293.5 of this group appears in releases 3.96 to 3.99
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 1-10 | 2-9 | 3-8 | 3-9 | 4-7 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HL_4WSM_251 | 4WSM | 0.7067 | 0 | 1K | tRNA | C | 47 | C | 48 | C | 49 | A | 50 | A | 51 | U | 52 | A | 53 | G | 54 | G | 55 | G | 56 | ncWW | ncWW | ntSH | ||
| 2 | HL_5J7L_212 | 5J7L | 0.5175 | 0 | DA | LSU rRNA | G | 1091 | C | 1092 | G | 1093 | U | 1094 | A | 1095 | A | 1096 | U | 1097 | A | 1098 | G | 1099 | C | 1100 | cWW | tSH | |||
| 3 | HL_4TUE_219 | 4TUE | 0.3400 | 0 | QY | A-site ASL-SufJ | C | 31 | C | 32 | U | 33 | U | 34 | G | 35 | G | 36 | U | 37 | A | 38 | A | 39 | G | 40 | cWW | ntWH | |||
| 4 | HL_8P9A_209 | 8P9A | 0.1493 | 0 | sR | SSU rRNA | A | 900 | G | 901 | G | 902 | U | 903 | G | 904 | A | 905 | A | 906 | A | 907 | U | 908 | U | 909 | cWW | ntSH | |||
| 5 | HL_8CRE_210 | 8CRE | 0.0000 | 0 | CM | SSU rRNA | A | 885 | G | 886 | G | 887 | U | 888 | G | 889 | A | 890 | A | 891 | A | 892 | U | 893 | U | 894 | cWW | tSH | ntSH | ||
| 6 | HL_4LFB_015 | 4LFB | 0.1177 | 0 | A | SSU rRNA | C | 689 | G | 690 | G | 691 | U | 692 | G | 693 | A | 694 | A | 695 | A | 696 | U | 697 | G | 698 | cWW | tSH | tSH | ||
| 7 | HL_5J7L_015 | 5J7L | 0.1126 | 0 | AA | SSU rRNA | C | 689 | G | 690 | G | 691 | U | 692 | G | 693 | A | 694 | A | 695 | A | 696 | U | 697 | G | 698 | cWW | tSH | tSH | ||
| 8 | HL_8P9A_211 | 8P9A | 0.3192 | 1 | sR | SSU rRNA | G | 997 | A | 998 | U | 999 | C | 1000 | A | 1001 | G | 1002 | A | 1003 | A | 1005 | C | 1006 | C | 1007 | cWW | ||||
| 9 | HL_4LFB_017 | 4LFB | 0.3394 | 1 | A | SSU rRNA | G | 786 | A | 787 | U | 788 | U | 789 | A | 790 | G | 791 | A | 792 | A | 794 | C | 795 | C | 796 | cWW | ncWB | |||
| 10 | HL_5J7L_017 | 5J7L | 0.3203 | 1 | AA | SSU rRNA | G | 786 | A | 787 | U | 788 | U | 789 | A | 790 | G | 791 | A | 792 | A | 794 | C | 795 | C | 796 | cWW | ncWB | |||
| 11 | HL_8CRE_212 | 8CRE | 0.3327 | 1 | CM | SSU rRNA | G | 982 | A | 983 | U | 984 | C | 985 | A | 986 | G | 987 | A | 988 | A | 990 | C | 991 | C | 992 | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| AGGUGAAAUU | 2 |
| CGGUGAAAUG | 2 |
| GAUCAGAUACC | 2 |
| GAUUAGAUACC | 2 |
| CCCAAUAGGG | 1 |
| GCGUAAUAGC | 1 |
| CCUUGGUAAG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GGUGAAAU | 4 |
| AUCAGAUAC | 2 |
| AUUAGAUAC | 2 |
| CCAAUAGG | 1 |
| CGUAAUAG | 1 |
| CUUGGUAA | 1 |
- Annotations
-
- (11)
- Basepair signature
- cWW-F-F-F-F-F-F-F-F
- Heat map statistics
- Min 0.07 | Avg 0.37 | Max 0.74
Coloring options: