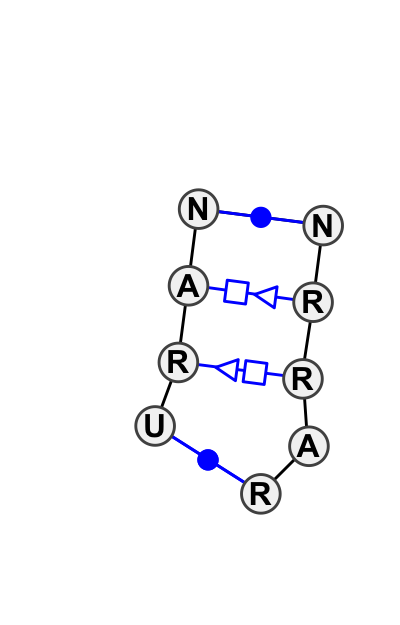
Motif IL_31555.7 Version IL_31555.7 of this group appears in releases 1.17 to 1.17
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | break | 5 | 6 | 7 | 8 | 9 | 1-9 | 2-7 | 3-6 | 4-5 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_1VX6_125 | 1VX6 | 0.5032 | 1 | A | LSU rRNA | U | 3440 | A | 3441 | A | 3443 | G | 3444 | * | C | 3469 | G | 3470 | A | 3471 | A | 3472 | G | 3473 | cWW | cWW | |||
| 2 | IL_4A1B_120 | 4A1B | 0.3655 | 2 | 1 | LSU rRNA | U | 3043 | A | 3044 | A | 3046 | A | 3048 | * | U | 3073 | G | 3074 | A | 3075 | A | 3076 | G | 3077 | cWW | tHS | cWW | ||
| 3 | IL_3J7A_006 | 3J7A | 0.4650 | 1 | A | SSU rRNA | U | 64 | A | 65 | A | 67 | U | 68 | * | A | 84 | G | 85 | A | 86 | A | 87 | A | 88 | cWW | tHS | cWW | ||
| 4 | IL_3U5F_007 | 3U5F | 0.4376 | 2 | 6 | SSU rRNA | U | 64 | A | 65 | A | 67 | G | 69 | * | U | 82 | G | 83 | A | 84 | A | 85 | A | 86 | cWW | tHS | cWW | ||
| 5 | IL_4CUX_007 | 4CUX | 0.4420 | 2 | 2 | SSU rRNA | U | 64 | A | 65 | A | 67 | G | 69 | * | U | 82 | G | 83 | A | 84 | A | 85 | A | 86 | cWW | ntHS | ncWW | ||
| 6 | IL_1GID_003 | 1GID | 0.3005 | 0 | A | P4-P6 RNA RIBOZYME DOMAIN | U | 205 | A | 206 | A | 207 | C | 208 | * | G | 112 | A | 113 | A | 114 | A | 115 | G | 116 | ncWW | tSH | tHS | cWW | |
| 7 | IL_1GID_010 | 1GID | 0.2771 | 0 | B | P4-P6 RNA RIBOZYME DOMAIN | U | 205 | A | 206 | A | 207 | C | 208 | * | G | 112 | A | 113 | A | 114 | A | 115 | G | 116 | cWW | tSH | tHS | cWW | |
| 8 | IL_1X8W_002 | 1X8W | 0.2701 | 0 | A | Group I catalytic intron | U | 205 | A | 206 | A | 207 | C | 208 | * | G | 112 | A | 113 | A | 114 | A | 115 | G | 116 | ncWW | tHS | cWW | ||
| 9 | IL_4A1B_022 | 4A1B | 0.2513 | 0 | 1 | LSU rRNA | U | 499 | G | 500 | A | 501 | G | 502 | * | C | 475 | A | 476 | G | 477 | A | 478 | G | 479 | cWW | tSH | ntHS | cWW | |
| 10 | IL_4A1B_026 | 4A1B | 0.1468 | 0 | 1 | LSU rRNA | U | 571 | G | 572 | A | 573 | G | 574 | * | C | 594 | A | 595 | A | 596 | A | 597 | A | 598 | cWW | ntSH | tHS | cWW | |
| 11 | IL_4FAW_010 | 4FAW | 0.0000 | 0 | A | Group II catalytic intron D1-D4-3 | U | 172 | G | 173 | A | 174 | G | 175 | * | C | 190 | A | 191 | A | 192 | A | 193 | G | 194 | cWW | tSH | tHS | cWW | |
| 12 | IL_3FTM_001 | 3FTM | 0.3121 | 0 | A+B | RNA (11-mer) + RNA (12-mer) | U | 46 | G | 47 | A | 48 | G | 49 | * | C | -1 | G | 1 | G | 2 | A | 3 | A | 4 | cWW | ntHS | cWW | ||
| 13 | IL_3FS0_001 | 3FS0 | 0.4073 | 0 | UAA/GAN related | A+B | RNA (10-mer) + RNA (11-mer) | U | 46 | G | 47 | A | 48 | G | 49 | * | C | -1 | G | 1 | G | 2 | A | 3 | A | 4 | cWW | nthH | tHS | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| UAAC*GAAAG | 3 |
| UAUAAG*UGAAA | 2 |
| UGAG*CGGAA | 2 |
| UACAG*CGAAG | 1 |
| UAAAUA*UGAAG | 1 |
| UAUAU*AGAAA | 1 |
| UGAG*CAGAG | 1 |
| UGAG*CAAAA | 1 |
| UGAG*CAAAG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| AA*AAA | 3 |
| AUAA*GAA | 2 |
| GA*AAA | 2 |
| GA*GGA | 2 |
| ACA*GAA | 1 |
| AAAU*GAA | 1 |
| AUA*GAA | 1 |
| GA*AGA | 1 |
Release history
| Release | 1.17 |
|---|---|
| Date | 2014-09-04 |
| Status | Updated, 2 parents |
Parent motifs
| Parent motif | Common motif instances | Only in IL_31555.7 | Only in the parent motif |
|---|---|---|---|
| IL_31555.6 Compare | IL_4A1B_026, IL_1GID_010, IL_1X8W_002, IL_4A1B_022, IL_3FS0_001, IL_3FTM_001, IL_4A1B_120, IL_1GID_003, IL_4FAW_010 | IL_3J7A_006, IL_3U5F_007, IL_4CUX_007, IL_1VX6_125 | IL_3LOA_001, IL_3BNT_005, IL_3BNQ_002, IL_3BNT_002 |
| IL_21427.1 Compare | IL_3U5F_007, IL_4CUX_007 | IL_1VX6_125, IL_4A1B_026, IL_1GID_010, IL_1X8W_002, IL_4A1B_022, IL_3FS0_001, IL_3FTM_001, IL_4A1B_120, IL_1GID_003, IL_4FAW_010, IL_3J7A_006 | IL_4CUV_104, IL_4BPP_007, IL_2QBG_075 |
Children motifs
| Child motif | Common motif instances | Only in IL_31555.7 | Only in the child motif |
|---|---|---|---|
| IL_31555.8 Compare | IL_1X8W_002, IL_1VX6_125, IL_4A1B_026, IL_3J7A_006, IL_3U5F_007, IL_4CUX_007, IL_4A1B_022, IL_3FS0_001, IL_1GID_003, IL_3FTM_001, IL_4A1B_120, IL_1GID_010, IL_4FAW_010 | IL_4W23_006, IL_4W21_004 |
- Annotations
-
- (12)
- UAA/GAN related (1)
- Basepair signature
- cWW-R-tSH-tHS-cWW
- Heat map statistics
- Min 0.15 | Avg 0.38 | Max 0.60
Coloring options: