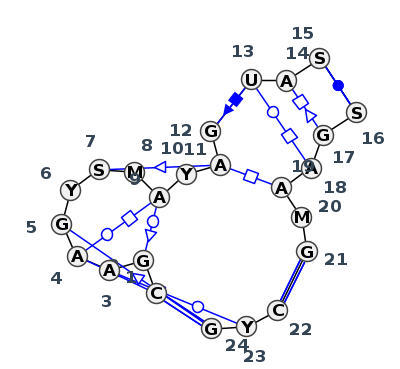
Motif J3_07616.1 Version J3_07616.1 of this group appears in releases 4.0 to 4.6
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | break | 16 | 17 | 18 | 19 | 20 | 21 | break | 22 | 23 | 24 | 1-4 | 1-24 | 2-9 | 3-23 | 4-9 | 5-24 | 7-11 | 11-19 | 12-13 | 13-18 | 14-17 | 15-16 | 21-22 | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | J3_4WF9_012 | 4WF9 | 0.2197 | 3 | X | LSU rRNA | C | 491 | G | 492 | A | 493 | A | 495 | G | 496 | U | 497 | G | 498 | A | 499 | A | 500 | C | 501 | A | 503 | G | 504 | U | 505 | A | 506 | C | 507 | * | G | 514 | G | 515 | A | 516 | A | 517 | A | 518 | G | 519 | * | C | 32 | U | 33 | G | 35 | cWW | tSW | tHW | tWH | ntSS | cSH | tWH | cWW | cWW | ||||
| 2 | J3_9DFE_003 | 9DFE | 0.1948 | 3 | 1A | LSU rRNA | C | 445 | G | 446 | A | 447 | A | 449 | G | 450 | C | 451 | G | 452 | C | 453 | A | 454 | C | 455 | A | 457 | G | 458 | U | 459 | A | 460 | C | 461 | * | G | 468 | G | 469 | A | 470 | A | 471 | A | 472 | G | 473 | * | C | 32 | U | 33 | G | 35 | ncSs | cWW | tSW | tHW | tWH | tSS | tsS | tHH | cSH | tWH | tHS | cWW | cWW |
| 3 | J3_4V9F_013 | 4V9F | 0.0878 | 3 | 0 | LSU rRNA | C | 451 | G | 452 | A | 453 | A | 455 | G | 456 | U | 457 | G | 458 | A | 459 | A | 460 | C | 461 | A | 463 | G | 464 | U | 465 | A | 466 | G | 467 | * | C | 474 | G | 475 | A | 476 | A | 477 | C | 478 | G | 479 | * | C | 29 | U | 30 | G | 32 | ncSs | cWW | tSW | tHW | tWH | tSS | tsS | tHH | cSH | tWH | tHS | cWW | cWW |
| 4 | J3_9E6Q_003 | 9E6Q | 0.0693 | 3 | 1 | LSU rRNA | C | 476 | G | 477 | A | 478 | A | 480 | G | 481 | C | 482 | G | 483 | C | 484 | A | 485 | C | 486 | A | 488 | G | 489 | U | 490 | A | 491 | OMC | 492 | * | G | 499 | G | 500 | A | 501 | A | 502 | A | 503 | G | 504 | * | C | 25 | U | 26 | G | 28 | cSs | cWW | tSW | tHW | tWH | tSS | tsS | tHH | cSH | tWH | tHS | cWW | cWW |
| 5 | J3_8OI5_004 | 8OI5 | 0.0458 | 3 | 1+4 | LSU rRNA + 5.8S rRNA | C | 340 | G | 341 | A | 342 | A | 344 | G | 345 | C | 346 | G | 347 | A | 348 | A | 349 | C | 350 | A | 352 | G | 353 | U | 354 | A | 355 | C | 356 | * | G | 363 | G | 364 | A | 365 | A | 366 | A | 367 | G | 368 | * | C | 21 | U | 22 | G | 24 | cSs | cWW | tSW | tHW | tWH | tSS | tsS | tHH | cSH | tWH | tHS | cWW | cWW |
| 6 | J3_8P9A_045 | 8P9A | 0.0448 | 3 | AR+AT | LSU rRNA + 5.8S rRNA | C | 340 | G | 341 | A | 342 | A | 344 | G | 345 | C | 346 | G | 347 | A | 348 | A | 349 | C | 350 | A | 352 | G | 353 | U | 354 | A | 355 | C | 356 | * | G | 363 | G | 364 | A | 365 | A | 366 | A | 367 | G | 368 | * | C | 21 | U | 22 | G | 24 | cSs | cWW | tSW | tHW | ntWH | tSS | tsS | tHH | cSH | tWH | tHS | cWW | cWW |
| 7 | J3_8GLP_024 | 8GLP | 0.0000 | 3 | L5+L8 | LSU rRNA + 5.8S rRNA | C | 351 | G | 352 | A | 353 | A | 355 | G | 356 | U | 357 | C | 358 | A | 359 | A | 360 | C | 361 | A | 363 | G | 364 | U | 365 | A | 366 | C | 367 | * | G | 374 | G | 375 | A | 376 | A | 377 | A | 378 | G | 379 | * | C | 21 | U | 22 | G | 24 | cSs | cWW | tSW | tHW | tWH | tSS | tsS | tHH | cSH | tWH | tHS | cWW | cWW |
| 8 | J3_9AXU_004 | 9AXU | 0.0410 | 3 | 2+4 | LSU rRNA + 5.8S rRNA | C | 348 | G | 349 | A | 350 | A | 352 | G | 353 | C | 354 | G | 355 | A | 356 | A | 357 | C | 358 | A | 360 | G | 361 | U | 362 | A | 363 | G | 364 | * | C | 371 | G | 372 | A | 373 | A | 374 | A | 375 | G | 376 | * | C | 29 | U | 30 | G | 32 | ncSs | cWW | tSW | tHW | tWH | tSS | tsS | tHH | cSH | tWH | tHS | cWW | cWW |
| 9 | J3_8B0X_033 | 8B0X | 0.0710 | 3 | a | LSU rRNA | C | 445 | G | 446 | A | 447 | A | 449 | G | 450 | U | 451 | G | 452 | A | 453 | A | 454 | C | 455 | A | 457 | G | 458 | U | 459 | A | 460 | C | 461 | * | G | 468 | G | 469 | A | 470 | A | 471 | A | 472 | G | 473 | * | C | 32 | C | 33 | G | 35 | ncSs | cWW | tSW | tHW | tWH | tSS | tsS | tHH | cSH | tWH | tHS | cWW | cWW |
| 10 | J3_7A0S_016 | 7A0S | 0.3693 | 2 | X | LSU rRNA | C | 457 | G | 458 | A | 459 | A | 461 | G | 462 | C | 463 | G | 464 | C | 465 | A | 466 | U | 467 | A | 468 | G | 469 | U | 470 | A | 471 | C | 472 | * | G | 479 | G | 480 | A | 481 | A | 482 | A | 483 | G | 484 | * | C | 32 | C | 33 | G | 35 | cSs | cWW | tSW | tHW | ntWH | tSS | tsS | tHH | cSH | tWH | tHS | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CGAUAGCGAACAAGUAC*GGAAAG*CUUG | 2 |
| CUUG*CGAUAGUGAACCAGUAC*GGAAAG | 1 |
| CUCG*CGAUAGCGCACCAGUAC*GGAAAG | 1 |
| CUCG*CGAUAGUGAACAAGUAG*CGAACG | 1 |
| CUCG*CGAUAGCGCACUAGUAOMC*GGAAAG | 1 |
| CGAUAGUCAACAAGUAC*GGAAAG*CUCG | 1 |
| CGAUAGCGAACAAGUAG*CGAAAG*CUUG | 1 |
| CCUG*CGAUAGUGAACCAGUAC*GGAAAG | 1 |
| CCUG*CGAUAGCGCAUAGUAC*GGAAAG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GAUAGCGAACAAGUA*GAAA*UU | 3 |
| UU*GAUAGUGAACCAGUA*GAAA | 1 |
| UC*GAUAGCGCACCAGUA*GAAA | 1 |
| UC*GAUAGUGAACAAGUA*GAAC | 1 |
| UC*GAUAGCGCACUAGUA*GAAA | 1 |
| GAUAGUCAACAAGUA*GAAA*UC | 1 |
| CU*GAUAGUGAACCAGUA*GAAA | 1 |
| CU*GAUAGCGCAUAGUA*GAAA | 1 |
Release history
| Release | 4.0 | 4.1 | 4.2 | 4.3 | 4.4 | 4.5 | 4.6 |
|---|---|---|---|---|---|---|---|
| Date | 2025-08-13 | 2025-09-10 | 2025-10-08 | 2025-11-05 | 2025-12-03 | 2025-12-31 | 2026-01-28 |
| Status | New id, 1 parent | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (10)
- Basepair signature
- cWW-cSS-tSS-tSW-tHW-cWW-tWH-F-F-tHH-tSS-tWH-F-tHS-cWW-F-cSH
- Heat map statistics
- Min 0.04 | Avg 0.16 | Max 0.40
Coloring options: