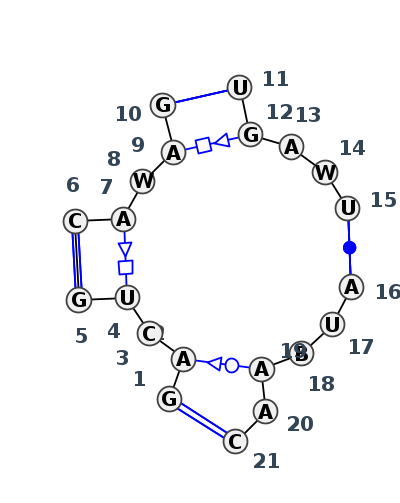
Motif J4_78485.1 Version J4_78485.1 of this group appears in releases 4.0 to 4.6
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | break | 6 | 7 | 8 | 9 | 10 | break | 11 | 12 | 13 | 14 | 15 | break | 16 | 17 | 18 | 19 | 20 | 21 | 1-18 | 1-21 | 2-19 | 3-8 | 4-7 | 5-6 | 9-12 | 10-11 | 13-18 | 15-16 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | J4_9E6Q_007 | 9E6Q | 0.2154 | 0 | 1 | LSU rRNA | G | 1908 | A | 1909 | C | 1910 | PSU | 1911 | G | 1912 | * | C | 1924 | A | 1925 | U | 1926 | A | 1927 | G | 1928 | * | U | 1963 | G | 1964 | A | 1965 | A | 1966 | U | 1967 | * | A | 2095 | U | 2096 | C | 2097 | A | 2098 | A | 2099 | C | 2100 | tHW | cWW | tSW | ntHS | ntHS | cWW | tHS | cWW | cWW | |
| 2 | J4_4V9F_007 | 4V9F | 0.0701 | 0 | 0 | LSU rRNA | G | 1828 | A | 1829 | C | 1830 | U | 1831 | G | 1832 | * | C | 1844 | A | 1845 | U | 1846 | A | 1847 | G | 1848 | * | U | 1883 | G | 1884 | A | 1885 | A | 1886 | U | 1887 | * | A | 2015 | U | 2016 | U | 2017 | A | 2018 | A | 2019 | C | 2020 | cWW | tSW | ntHSa | ntHS | cWW | tHS | cWW | cWW | ||
| 3 | J4_9AXU_007 | 9AXU | 0.0000 | 0 | 2 | LSU rRNA | G | 2218 | A | 2219 | C | 2220 | U | 2221 | G | 2222 | * | C | 2234 | A | 2235 | U | 2236 | A | 2237 | G | 2238 | * | U | 2274 | G | 2275 | A | 2276 | U | 2277 | U | 2278 | * | A | 2405 | U | 2406 | U | 2407 | A | 2408 | A | 2409 | C | 2410 | cWW | tSW | ntHS | tHS | cWW | tHS | cWW | cWW | ||
| 4 | J4_8P9A_017 | 8P9A | 0.0411 | 0 | AR | LSU rRNA | G | 2130 | A | 2131 | C | 2132 | U | 2133 | G | 2134 | * | C | 2146 | A | 2147 | U | 2148 | A | 2149 | G | 2150 | * | U | 2186 | G | 2187 | A | 2188 | U | 2189 | U | 2190 | * | A | 2317 | U | 2318 | U | 2319 | A | 2320 | A | 2321 | C | 2322 | cWW | tSW | ntHSa | tHS | cWW | tHS | cWW | ntHS | cWW | |
| 5 | J4_8OI5_006 | 8OI5 | 0.0503 | 0 | 1 | LSU rRNA | G | 2108 | A | 2109 | C | 2110 | U | 2111 | G | 2112 | * | C | 2124 | A | 2125 | U | 2126 | A | 2127 | G | 2128 | * | U | 2164 | G | 2165 | A | 2166 | U | 2167 | U | 2168 | * | A | 2295 | U | 2296 | U | 2297 | A | 2298 | A | 2299 | C | 2300 | cWW | tSW | ntHSa | tHS | cWW | tHS | cWW | ntHS | cWW | |
| 6 | J4_8GLP_009 | 8GLP | 0.1334 | 0 | L5 | LSU rRNA | G | 3634 | A | 3635 | C | 3636 | PSU | 3637 | G | 3638 | * | C | 3650 | A | 3651 | A | 3652 | A | 3653 | G | 3654 | * | U | 3690 | G | 3691 | A | 3692 | U | 3693 | U | 3694 | * | A | 3821 | U | 3822 | G | 3823 | A | 3824 | A2M | 3825 | C | 3826 | cWW | tSW | ntHS | tHS | cWW | tHS | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GACUG*CAUAG*UGAUU*AUUAAC | 3 |
| GAC(PSU)G*CAUAG*UGAAU*AUCAAC | 1 |
| GACUG*CAUAG*UGAAU*AUUAAC | 1 |
| GAC(PSU)G*CAAAG*UGAUU*AUGA(A2M)C | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| ACU*AUA*GAU*UUAA | 3 |
| AC(PSU)*AUA*GAA*UCAA | 1 |
| ACU*AUA*GAA*UUAA | 1 |
| AC(PSU)*AAA*GAU*UGA(A2M) | 1 |
Release history
| Release | 4.0 | 4.1 | 4.2 | 4.3 | 4.4 | 4.5 | 4.6 |
|---|---|---|---|---|---|---|---|
| Date | 2025-08-13 | 2025-09-10 | 2025-10-08 | 2025-11-05 | 2025-12-03 | 2025-12-31 | 2026-01-28 |
| Status | New id, 1 parent | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (6)
- Basepair signature
- cWW-tSW-F-F-tHS-F-cWW-F-cWW-F-F-tHS-F-cWW
- Heat map statistics
- Min 0.04 | Avg 0.12 | Max 0.26
Coloring options: