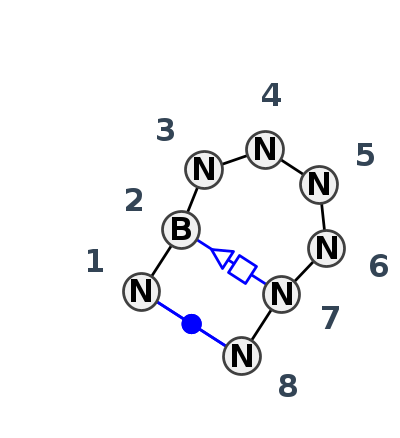
Motif HL_84085.4 Version HL_84085.4 of this group appears in releases 3.88 to 3.88
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 1-8 | 2-7 | 3-6 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HL_5J7L_012 | 5J7L | 0.4101 | 1 | GNRA wlth extra near cWW | AA | SSU rRNA | G | 462 | U | 463 | U | 464 | A | 465 | A | 466 | A | 468 | C | 469 | C | 470 | cWW | ||
| 2 | HL_4V9F_043 | 4V9F | 0.3959 | 1 | GNRA wlth tandem sheared | 0 | LSU rRNA | C | 1705 | G | 1706 | G | 1707 | C | 1708 | G | 1709 | A | 1711 | A | 1712 | G | 1713 | cWW | tSH | ntSH |
| 3 | HL_3HHN_010 | 3HHN | 0.2474 | 0 | GNRA wlth tandem sheared | E | Class I ligase ribozyme, self-ligation product | G | 92 | U | 93 | U | 94 | A | 95 | A | 96 | A | 97 | A | 98 | C | 99 | cWW | ntSH | |
| 4 | HL_8VTW_021 | 8VTW | 0.2701 | 0 | GNRA wlth tandem sheared | 1A | LSU rRNA | G | 712 | G | 713 | U | 714 | G | 715 | A | 716 | G | 717 | A | 718 | C | 719 | cWW | ntSH | |
| 5 | HL_4V9F_049 | 4V9F | 0.2308 | 0 | GNRA related | 0 | LSU rRNA | C | 1916 | G | 1917 | U | 1918 | A | 1919 | C | 1920 | A | 1921 | A | 1922 | G | 1923 | cWW | ntSH | |
| 6 | HL_5J7L_154 | 5J7L | 0.0000 | 0 | GNRA wlth tandem sheared | DA | LSU rRNA | G | 712 | G | 713 | U | 714 | A | 715 | A | 716 | C | 717 | A | 718 | C | 719 | cWW | tSH | |
| 7 | HL_1MMS_002 | 1MMS | 0.1661 | 0 | GNRA wlth tandem sheared | C | LSU rRNA | C | 1092 | G | 1093 | U | 1094 | A | 1095 | A | 1096 | C | 1097 | A | 1098 | G | 1099 | cWW | tSH | ntSHa |
| 8 | HL_6PRV_002 | 6PRV | 0.1693 | 0 | GNRA wlth tandem sheared | A | LSU rRNA | C | 1092 | G | 1093 | U | 1094 | A | 1095 | A | 1096 | U | 1097 | A | 1098 | G | 1099 | cWW | tSH | |
| 9 | HL_5D8H_002 | 5D8H | 0.1609 | 0 | GNRA wlth tandem sheared | A | LSU rRNA | C | 1202 | G | 1203 | U | 1204 | A | 1205 | A | 1206 | C | 1207 | A | 1208 | G | 1209 | cWW | tSH | |
| 10 | HL_8C3A_028 | 8C3A | 0.1697 | 0 | GNRA wlth tandem sheared | 1 | LSU rRNA | U | 1263 | G | 1264 | U | 1265 | A | 1266 | A | 1267 | C | 1268 | A | 1269 | A | 1270 | cWW | tSH | |
| 11 | HL_4WF9_004 | 4WF9 | 0.2873 | 0 | GNRA wlth tandem sheared | X | LSU rRNA | A | 136 | G | 137 | U | 138 | U | 139 | A | 140 | U | 141 | G | 142 | U | 143 | cWW | ||
| 12 | HL_5AOX_001 | 5AOX | 0.2584 | 0 | GNRA wlth tandem sheared | C | ALU JO CONSENSUS RNA | G | 11 | G | 12 | U | 13 | G | 14 | G | 15 | C | 16 | U | 17 | C | 18 | cWW | ntSH | |
| 13 | HL_1E8O_001 | 1E8O | 0.2889 | 0 | GNRA wlth tandem sheared | E | 7SL RNA | G | 110 | G | 111 | U | 112 | G | 113 | G | 114 | C | 115 | G | 116 | C | 117 | cWW | ntSH | ntSHa |
| 14 | HL_4KQY_003 | 4KQY | 0.3723 | 1 | GNRA wlth tandem sheared | A | SAM-I riboswitch | C | 94 | U | 95 | C | 96 | G | 97 | A | 98 | A | 99 | C | 100 | G | 102 | cWW | ||
| 15 | HL_2IL9_007 | 2IL9 | 0.4466 | 0 | GNRA with extra near cWW | A | CrPV IRES | G | 6118 | C | 6119 | G | 6119|||A | A | 6119|||B | A | 6119|||C | A | 6119|||D | G | 6119|||E | C | 6124 | cWW | ncWW | tSH |
| 16 | HL_6CZR_215 | 6CZR | 0.5972 | 0 | GNRA with extra near cWW | 1a | SSU rRNA | G | 152 | G | 153 | G | 154 | A | 155 | A | 156 | A | 157 | C | 158 | U | 159 | ncWW | ncWW | ntSH |
| 17 | HL_4V87_253 | 4V87 | 0.5880 | 0 | GNRA with extra near cWW | BB | tRNA | C | 48 | C | 49 | A | 50 | A | 51 | U | 52 | A | 53 | G | 54 | G | 55 | ncWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CGUAACAG | 2 |
| GUUAAUACC | 1 |
| CGGCGAAAG | 1 |
| GUUAAAAC | 1 |
| GGUGAGAC | 1 |
| CGUACAAG | 1 |
| GGUAACAC | 1 |
| CGUAAUAG | 1 |
| UGUAACAA | 1 |
| AGUUAUGU | 1 |
| GGUGGCUC | 1 |
| GGUGGCGC | 1 |
| CUCGAACAG | 1 |
| GCGAAAGC | 1 |
| GGGAAACU | 1 |
| CCAAUAGG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GUAACA | 4 |
| UUAAUAC | 1 |
| GGCGAAA | 1 |
| UUAAAA | 1 |
| GUGAGA | 1 |
| GUACAA | 1 |
| GUAAUA | 1 |
| GUUAUG | 1 |
| GUGGCU | 1 |
| GUGGCG | 1 |
| UCGAACA | 1 |
| CGAAAG | 1 |
| GGAAAC | 1 |
| CAAUAG | 1 |
Release history
| Release | 3.88 |
|---|---|
| Date | 2024-09-11 |
| Status | Updated, 1 parent |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- GNRA wlth tandem sheared (12)
- GNRA with extra near cWW (3)
- GNRA related (1)
- GNRA wlth extra near cWW (1)
- Basepair signature
- cWW-tSH-F-F-F-F
- Heat map statistics
- Min 0.08 | Avg 0.40 | Max 0.82
Coloring options: