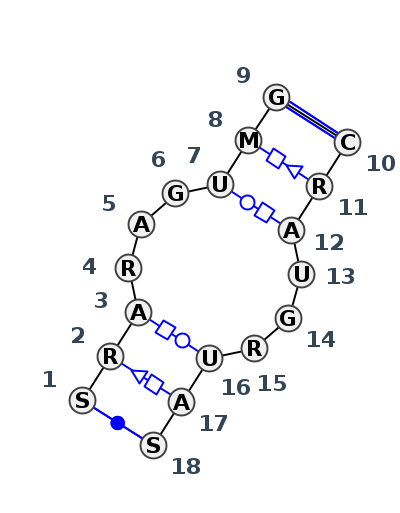
Motif IL_56455.5 Version IL_56455.5 of this group appears in releases 3.88 to 3.90
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | break | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 1-18 | 2-17 | 3-16 | 4-15 | 6-13 | 7-12 | 8-11 | 9-10 | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_364D_003 | 364D | 0.2030 | 0 | Bacterial 5S Loop E | C+B | RNA (31-mer) | C | 97 | G | 98 | A | 99 | G | 100 | A | 101 | G | 102 | U | 103 | A | 104 | G | 105 | * | C | 71 | G | 72 | A | 73 | U | 74 | G | 75 | G | 76 | U | 77 | A | 78 | G | 79 | cWW | tSH | ntHW | cBW | ntWH | tWH | tHS | cWW |
| 2 | IL_1FEU_001 | 1FEU | 0.1375 | 0 | Bacterial 5S Loop E | C+B | 21 NT FRAGMENT OF 5S RRNA | C | 97 | G | 98 | A | 99 | G | 100 | A | 101 | G | 102 | U | 103 | A | 104 | G | 105 | * | C | 71 | G | 72 | A | 73 | U | 74 | G | 75 | G | 76 | U | 77 | A | 78 | G | 79 | cWW | tSH | tHW | cBW | ncWB | tWH | tHS | cWW |
| 3 | IL_354D_001 | 354D | 0.0590 | 0 | Bacterial 5S Loop E | B+A | RNA-DNA (5'-R(*GP*CP*GP*AP*GP*AP*GP*UP*AP*)-D(*DGP(S)*)-R(*GP*C)-3') | C | 97 | G | 98 | A | 99 | G | 100 | A | 101 | G | 102 | U | 103 | A | 104 | DG | 105 | * | C | 71 | G | 72 | A | 73 | U | 74 | G | 75 | G | 76 | U | 77 | A | 78 | G | 79 | cWW | tSH | tHW | cBW | cWB | tWH | tHS | cWW |
| 4 | IL_1DFU_001 | 1DFU | 0.0000 | 0 | Bacterial 5S Loop E | M+N | 5S RRNA | C | 97 | G | 98 | A | 99 | G | 100 | A | 101 | G | 102 | U | 103 | A | 104 | G | 105 | * | C | 71 | G | 72 | A | 73 | U | 74 | G | 75 | G | 76 | U | 77 | A | 78 | G | 79 | cWW | tSH | tHW | cBW | cWB | tWH | tHS | cWW |
| 5 | IL_5J7L_359 | 5J7L | 0.1247 | 0 | Bacterial 5S Loop E | DB | 5S rRNA | C | 97 | G | 98 | A | 99 | G | 100 | A | 101 | G | 102 | U | 103 | A | 104 | G | 105 | * | C | 71 | G | 72 | A | 73 | U | 74 | G | 75 | G | 76 | U | 77 | A | 78 | G | 79 | cWW | tSH | tHW | cBW | cWB | tWH | tHS | cWW |
| 6 | IL_8VTW_114 | 8VTW | 0.1332 | 0 | Bacterial 5S Loop E | 1B | 5S rRNA | G | 98 | G | 99 | A | 100 | G | 101 | A | 102 | G | 103 | U | 104 | A | 105 | G | 106 | * | C | 71 | G | 72 | A | 73 | U | 74 | G | 75 | G | 76 | U | 77 | A | 78 | C | 79 | cWW | tSH | tHW | cBW | ncWB | tWH | tHS | cWW |
| 7 | IL_7A0S_106 | 7A0S | 0.1589 | 0 | Bacterial 5S Loop E | Y | 5S rRNA | G | 100 | A | 101 | A | 102 | A | 103 | A | 104 | G | 105 | U | 106 | C | 107 | G | 108 | * | C | 73 | A | 74 | A | 75 | U | 76 | G | 77 | A | 78 | U | 79 | A | 80 | C | 81 | cWW | tSH | tHW | cBW | ncWB | tWH | tHS | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CGAGAGUAG*CGAUGGUAG | 5 |
| GGAGAGUAG*CGAUGGUAC | 1 |
| GAAAAGUCG*CAAUGAUAC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GAGAGUA*GAUGGUA | 6 |
| AAAAGUC*AAUGAUA | 1 |
- Annotations
-
- Bacterial 5S Loop E (7)
- Basepair signature
- cWW-tSH-tHW-L-R-L-R-L-R-tWH-tHS-cWW
- Heat map statistics
- Min 0.06 | Avg 0.14 | Max 0.24
Coloring options: