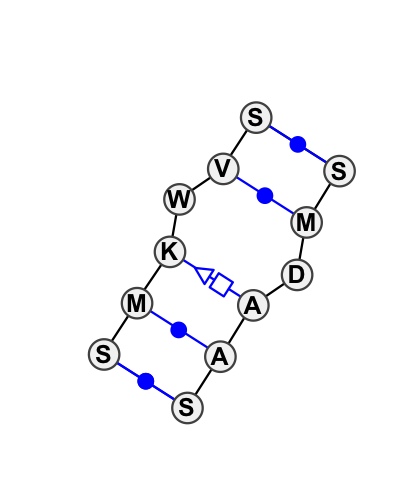
Motif IL_37171.1 Version IL_37171.1 of this group appears in releases 1.14 to 1.15
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | break | 7 | 8 | 9 | 10 | 11 | 12 | 1-12 | 2-10 | 2-11 | 3-4 | 3-9 | 3-10 | 4-9 | 5-8 | 6-7 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_3B5F_001 | 3B5F | 0.8258 | 0 | A+B | Loop A Substrate strand + 29-mer Loop A and Loop B Ribozyme strand | C | 4 | A | 5 | G | 6 | U | 7 | C | 8 | C | 9 | * | G | 6 | A | 7 | G | 8 | A | 9 | A | 10 | G | 11 | cWW | tSH | tHS | ncWWa | cWW | ||||
| 2 | IL_4NVV_033 | 4NVV | 0.0000 | 0 | A | LSU rRNA | G | 922 | C | 923 | U | 924 | A | 925 | G | 926 | G | 927 | * | C | 943 | C | 944 | A | 945 | A | 946 | A | 947 | C | 948 | cWW | cWW | ncSs | ncWW | tSH | ncWW | |||
| 3 | IL_3U5F_038 | 3U5F | 0.4939 | 0 | 6 | SSU rRNA | G | 730 | C | 731 | G | 732 | A | 733 | A | 734 | C | 735 | * | G | 703 | C | 704 | U | 705 | A | 706 | A | 707 | C | 708 | cWW | cWW | ntSH | ncWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CAGUCC*GAGAAG | 1 |
| GCUAGG*CCAAAC | 1 |
| GCGAAC*GCUAAC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| AGUC*AGAA | 1 |
| CUAG*CAAA | 1 |
| CGAA*CUAA | 1 |
Release history
Parent motifs
| Parent motif | Common motif instances | Only in IL_37171.1 | Only in the parent motif |
|---|---|---|---|
| IL_83856.1 Compare | IL_3U5F_038 | IL_4NVV_033, IL_3B5F_001 | |
| IL_80494.2 Compare | IL_3B5F_001 | IL_4NVV_033, IL_3U5F_038 | IL_3P49_012, IL_1S72_057 |
Children motifs
| Child motif | Common motif instances | Only in IL_37171.1 | Only in the child motif |
|---|---|---|---|
| IL_44977.1 Compare | IL_3U5F_038 | IL_4NVV_033, IL_3B5F_001 | IL_4A1B_025 |
| IL_26321.1 Compare | IL_3B5F_001 | IL_4NVV_033, IL_3U5F_038 | IL_4QK9_001, IL_4QK8_001, IL_4QLM_002 |
- Annotations
-
- (3)
- Basepair signature
- cWW-cWW-tSH-L-R-cWW-cWW
- Heat map statistics
- Min 0.49 | Avg 0.52 | Max 1.04
Coloring options: