Motif IL_70923.4 Version IL_70923.4 of this group appears in releases 3.88 to 3.88
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | break | 8 | 9 | 10 | 11 | 12 | 1-12 | 2-11 | 4-11 | 5-10 | 6-9 | 7-8 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
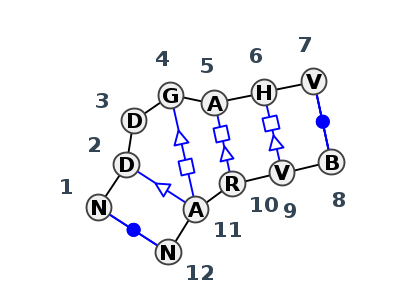
| 1 | IL_5J7L_330 | 5J7L | 0.4565 | 1 | Kink-turn | DA | LSU rRNA | C | 2129 | U | 2130 | U | 2131 | G | 2133 | A | 2134 | A | 2135 | G | 2136 | * | U | 2155 | G | 2156 | G | 2157 | A | 2158 | G | 2159 | cWW | ncSW | cWW | |||
| 2 | IL_7JRS_003 | 7JRS | 0.1960 | 1 | Kink-turn | B | RNA 3D nanocage | C | 12 | G | 13 | A | 14 | G | 16 | A | 17 | A | 18 | C | 19 | * | G | 120 | G | 121 | G | 122 | A | 123 | G | 124 | cWW | ntsS | tHS | tHS | cWW | |
| 3 | IL_4KQY_002 | 4KQY | 0.2045 | 1 | Kink-turn | A | SAM-I riboswitch | C | 31 | G | 32 | A | 33 | G | 35 | A | 36 | A | 37 | G | 38 | * | C | 17 | A | 18 | G | 19 | A | 20 | G | 21 | cWW | tsS | ntSH | ntHS | cWW | |
| 4 | IL_5Y7M_008 | 5Y7M | 0.1682 | 1 | Kink-turn | D | RNA (52-MER) | A | 17 | G | 18 | A | 19 | G | 21 | A | 22 | U | 23 | C | 24 | * | G | 29 | G | 30 | G | 31 | A | 32 | U | 33 | cWW | tsS | tSH | ntHS | ntHS | cWW |
| 5 | IL_7JRS_002 | 7JRS | 0.1419 | 1 | Kink-turn | A | RNA 3D nanocage | C | 77 | G | 78 | A | 79 | G | 81 | A | 82 | A | 83 | C | 84 | * | G | 55 | G | 56 | G | 57 | A | 58 | G | 59 | cWW | ntsS | tSH | tHS | tHS | cWW |
| 6 | IL_7JRR_001 | 7JRR | 0.1435 | 1 | Kink-turn | A | RNA (50-MER) | C | 23 | G | 24 | A | 25 | G | 27 | A | 28 | A | 29 | C | 30 | * | G | 5 | G | 6 | G | 7 | A | 8 | G | 9 | cWW | ntsS | tSH | tHS | tHS | cWW |
| 7 | IL_5FJ4_002 | 5FJ4 | 0.1475 | 1 | Kink-turn | H | HMKT-7 | C | 26 | G | 27 | A | 28 | G | 30 | A | 31 | A | 32 | C | 33 | * | G | 3 | G | 4 | G | 5 | A | 6 | G | 7 | cWW | ntsS | tSH | tHS | tHS | cWW |
| 8 | IL_4BW0_001 | 4BW0 | 0.1879 | 1 | Kink-turn | A | HMKT-7 | C | 16 | G | 17 | A | 18 | G | 20 | A | 21 | A | 22 | C | 23 | * | G | 3 | G | 4 | G | 5 | A | 6 | G | 7 | cWW | ntsS | tSH | tHS | tHS | cWW |
| 9 | IL_4C4W_002 | 4C4W | 0.2053 | 1 | Kink-turn | H | TSKT-23 | U | 26 | G | 27 | A | 28 | G | 30 | A | 31 | C | 32 | C | 33 | * | G | 3 | C | 4 | A | 5 | A | 6 | G | 7 | cWW | ntsS | tSH | tHS | ncBW | cWW |
| 10 | IL_7JRS_006 | 7JRS | 0.1717 | 1 | Kink-turn | B | RNA 3D nanocage | C | 77 | G | 78 | A | 79 | G | 81 | A | 82 | A | 83 | C | 84 | * | G | 55 | G | 56 | G | 57 | A | 58 | G | 59 | ncWW | ntsS | tSH | tHS | tHS | cWW |
| 11 | IL_7JRS_007 | 7JRS | 0.1599 | 1 | Kink-turn | A | RNA 3D nanocage | C | 12 | G | 13 | A | 14 | G | 16 | A | 17 | A | 18 | C | 19 | * | G | 120 | G | 121 | G | 122 | A | 123 | G | 124 | cWW | ntsS | tSH | tHS | tHS | cWW |
| 12 | IL_7EFG_001 | 7EFG | 0.0882 | 1 | Kink-turn | A | RNA (19-mer) | C | 3 | G | 4 | A | 5 | G | 7 | A | 8 | A | 9 | C | 10 | * | G | 13||||9_767 | G | 14||||9_767 | G | 15||||9_767 | A | 16||||9_767 | G | 17||||9_767 | cWW | ntsS | tSH | tHS | tHS | cWW |
| 13 | IL_7EFG_002 | 7EFG | 0.0882 | 1 | Kink-turn | A | RNA (19-mer) | C | 3||||9_767 | G | 4||||9_767 | A | 5||||9_767 | G | 7||||9_767 | A | 8||||9_767 | A | 9||||9_767 | C | 10||||9_767 | * | G | 13 | G | 14 | G | 15 | A | 16 | G | 17 | cWW | ntsS | tSH | tHS | tHS | cWW |
| 14 | IL_4CS1_002 | 4CS1 | 0.0885 | 1 | Kink-turn | A | 5'-(*GP*GP*CP*GP*AP*AP*GP*AP*AP*CP*CP*GP*GP*GP *GP*AP*GP*CP*CP)-3' | C | 3||||9_769 | G | 4||||9_769 | A | 5||||9_769 | G | 7||||9_769 | A | 8||||9_769 | A | 9||||9_769 | C | 10||||9_769 | * | G | 13 | G | 14 | G | 15 | A | 16 | G | 17 | cWW | ntsS | tSH | tHS | tHS | cWW |
| 15 | IL_4CS1_001 | 4CS1 | 0.0885 | 1 | Kink-turn | A | 5'-(*GP*GP*CP*GP*AP*AP*GP*AP*AP*CP*CP*GP*GP*GP *GP*AP*GP*CP*CP)-3' | C | 3 | G | 4 | A | 5 | G | 7 | A | 8 | A | 9 | C | 10 | * | G | 13||||9_769 | G | 14||||9_769 | G | 15||||9_769 | A | 16||||9_769 | G | 17||||9_769 | cWW | ntsS | tSH | tHS | tHS | cWW |
| 16 | IL_5G4T_002 | 5G4T | 0.0000 | 1 | Kink-turn | A+B | HMKT-7 | C | 3 | G | 4 | A | 5 | G | 7 | A | 8 | A | 9 | C | 10 | * | G | 13 | G | 14 | G | 15 | A | 16 | G | 17 | cWW | ntsS | tSH | tHS | tHS | cWW |
| 17 | IL_5FJC_002 | 5FJC | 0.1170 | 1 | Kink-turn | A | SAM-I riboswitch | C | 31 | G | 32 | A | 33||A | G | 35 | A | 36 | A | 37 | C | 38 | * | G | 17 | G | 18 | G | 19 | A | 20 | G | 21 | cWW | tsS | tSH | tHS | tHS | cWW |
| 18 | IL_3U4M_004 | 3U4M | 0.1365 | 1 | Kink-turn | B | LSU rRNA | C | 2129 | U | 2130 | G | 2131 | G | 2133 | A | 2134 | A | 2135 | C | 2136 | * | G | 2155 | G | 2156 | G | 2157 | A | 2158 | G | 2159 | cWW | tsS | tSH | tHS | tHS | cWW |
| 19 | IL_8VTW_046 | 8VTW | 0.1289 | 1 | Kink-turn | 1A | LSU rRNA | C | 1208 | G | 1209 | A | 1210 | G | 1212 | A | 1213 | A | 1214 | G | 1215 | * | U | 1234 | G | 1235 | G | 1236 | A | 1237 | G | 1238 | cWW | ntsS | tSH | tHS | tHS | cWW |
| 20 | IL_5J7L_288 | 5J7L | 0.1352 | 1 | Kink-turn | DA | LSU rRNA | C | 1208 | U | 1209 | G | 1210 | G | 1212 | A | 1213 | A | 1214 | G | 1215 | * | U | 1234 | G | 1235 | G | 1236 | A | 1237 | G | 1238 | cWW | tsS | tSH | tHS | tHS | cWW |
| 21 | IL_4V9F_048 | 4V9F | 0.1499 | 1 | Kink-turn | 0 | LSU rRNA | G | 1312 | A | 1313 | U | 1314 | G | 1316 | A | 1317 | A | 1318 | G | 1319 | * | U | 1338 | G | 1339 | G | 1340 | A | 1341 | C | 1342 | cWW | ntsS | tSH | tHS | tHS | cWW |
| 22 | IL_7EAG_001 | 7EAG | 0.1584 | 1 | Kink-turn | A+B | RNA (19-mer) | C | 3 | U | 4 | A | 5 | G | 7 | A | 8 | A | 9 | G | 10 | * | U | 13 | G | 14 | G | 15 | A | 16 | G | 17 | cWW | tsS | tSH | tHS | tHS | cWW |
| 23 | IL_7EAG_002 | 7EAG | 0.1738 | 1 | Kink-turn | C+D | RNA (19-mer) | C | 3 | U | 4 | A | 5 | G | 7 | A | 8 | A | 9 | G | 10 | * | U | 13 | G | 14 | G | 15 | A | 16 | G | 17 | cWW | ntsS | tSH | tHS | tHS | cWW |
| 24 | IL_7EAG_004 | 7EAG | 0.1663 | 1 | Kink-turn | E+F | RNA (19-mer) | C | 3 | U | 4 | A | 5 | G | 7 | A | 8 | A | 9 | G | 10 | * | U | 13 | G | 14 | G | 15 | A | 16 | G | 17 | cWW | ntsS | ntSH | tHS | tHS | cWW |
| 25 | IL_7EAG_003 | 7EAG | 0.1587 | 1 | Kink-turn | D+C | RNA (19-mer) | C | 3 | U | 4 | A | 5 | G | 7 | A | 8 | A | 9 | G | 10 | * | U | 13 | G | 14 | G | 15 | A | 16 | G | 17 | cWW | ntSH | ntHS | tHS | cWW | |
| 26 | IL_7EAG_005 | 7EAG | 0.1530 | 1 | Kink-turn | F+E | RNA (19-mer) | C | 3 | U | 4 | A | 5 | G | 7 | A | 8 | A | 9 | G | 10 | * | U | 13 | G | 14 | G | 15 | A | 16 | G | 17 | cWW | ntSH | ntHS | tHS | cWW | |
| 27 | IL_3V7E_002 | 3V7E | 0.1610 | 1 | Kink-turn | C | SAM-I riboswitch | C | 31 | G | 32 | A | 33 | G | 35 | A | 36 | A | 37 | A | 38 | * | U | 17 | G | 18 | G | 19 | A | 20 | G | 21 | cWW | tsS | tSH | tHS | cWW | |
| 28 | IL_5TBW_059 | 5TBW | 0.2264 | 1 | Kink-turn | 1 | LSU rRNA | U | 1388 | G | 1389 | A | 1390 | G | 1392 | A | 1393 | A | 1394 | G | 1395 | * | U | 1415 | C | 1416 | G | 1417 | A | 1418 | A | 1419 | cWW | tsS | tSH | tHS | tHS | cWW |
| 29 | IL_8C3A_063 | 8C3A | 0.2290 | 1 | Kink-turn | 1 | LSU rRNA | U | 1384 | G | 1385 | A | 1386 | G | 1388 | A | 1389 | A | 1390 | G | 1391 | * | U | 1411 | C | 1412 | G | 1413 | A | 1414 | A | 1415 | cWW | ntsS | tSH | tHS | tHS | cWW |
| 30 | IL_7A0S_043 | 7A0S | 0.1904 | 1 | Kink-turn | X | LSU rRNA | C | 1221 | G | 1222 | G | 1223 | G | 1225 | A | 1226 | A | 1227 | G | 1228 | * | U | 1247 | G | 1248 | G | 1249 | A | 1250 | G | 1251 | cWW | ntsS | tSH | ntHS | tHS | cWW |
| 31 | IL_4WF9_046 | 4WF9 | 0.1907 | 1 | Kink-turn | X | LSU rRNA | C | 1246 | G | 1247 | U | 1248 | G | 1250 | A | 1251 | A | 1252 | G | 1253 | * | U | 1272 | G | 1273 | G | 1274 | A | 1275 | G | 1276 | cWW | ntHS | tHS | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CGAAGAAC*GGGAG | 13 |
| CUAUGAAG*UGGAG | 5 |
| UGACGAAG*UCGAA | 2 |
| CUUUGAAG*UGGAG | 1 |
| CGACGAAG*CAGAG | 1 |
| AGACGAUC*GGGAU | 1 |
| UGAUGACC*GCAAG | 1 |
| CUGUGAAC*GGGAG | 1 |
| CGAUGAAG*UGGAG | 1 |
| CUGCGAAG*UGGAG | 1 |
| GAUGGAAG*UGGAC | 1 |
| CGAUGAAA*UGGAG | 1 |
| CGGAGAAG*UGGAG | 1 |
| CGUUGAAG*UGGAG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GAAGAA*GGA | 13 |
| UAUGAA*GGA | 5 |
| GAUGAA*GGA | 2 |
| GACGAA*CGA | 2 |
| UUUGAA*GGA | 1 |
| GACGAA*AGA | 1 |
| GACGAU*GGA | 1 |
| GAUGAC*CAA | 1 |
| UGUGAA*GGA | 1 |
| UGCGAA*GGA | 1 |
| AUGGAA*GGA | 1 |
| GGAGAA*GGA | 1 |
| GUUGAA*GGA | 1 |
Release history
| Release | 3.88 |
|---|---|
| Date | 2024-09-11 |
| Status | Updated, 1 parent |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- Kink-turn (31)
- Basepair signature
- cWW-tSS-tSH-L-tHS-tHS-cWW
- Heat map statistics
- Min 0.00 | Avg 0.21 | Max 0.54
Coloring options: